Project
DANIO-CODE
Navigation
Downloads
Results for rfx5
Z-value: 0.43
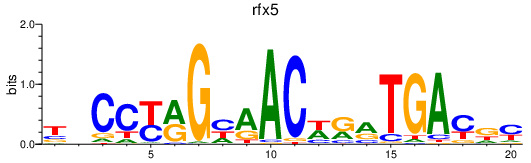
Transcription factors associated with rfx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rfx5
|
ENSDARG00000063258 | regulatory factor X, 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rfx5 | dr10_dc_chr19_-_7576069_7576133 | -0.77 | 5.2e-04 | Click! |
Activity profile of rfx5 motif
Sorted Z-values of rfx5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of rfx5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_28004606 | 1.51 |
ENSDART00000121643
|
si:dkeyp-46h3.2
|
si:dkeyp-46h3.2 |
| chr13_+_22691356 | 1.08 |
ENSDART00000144094
|
dna2
|
DNA replication helicase/nuclease 2 |
| chr10_-_36149719 | 0.90 |
|
|
|
| chr13_+_22691251 | 0.80 |
ENSDART00000144094
|
dna2
|
DNA replication helicase/nuclease 2 |
| chr12_+_14046373 | 0.73 |
|
|
|
| chr17_+_15527923 | 0.58 |
ENSDART00000148443
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr18_-_3455211 | 0.53 |
ENSDART00000163762
|
dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr13_+_22691194 | 0.52 |
ENSDART00000109798
|
dna2
|
DNA replication helicase/nuclease 2 |
| chr13_-_17729510 | 0.51 |
ENSDART00000170583
|
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr13_+_31586338 | 0.51 |
ENSDART00000034745
|
prkcha
|
protein kinase C, eta, a |
| chr13_+_16389616 | 0.49 |
|
|
|
| chr13_+_16389663 | 0.47 |
|
|
|
| chr20_-_29961589 | 0.47 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr1_+_5527701 | 0.47 |
ENSDART00000145378
|
fastkd2
|
FAST kinase domains 2 |
| chr6_-_25065823 | 0.45 |
ENSDART00000167259
|
znf326
|
zinc finger protein 326 |
| chr1_+_499061 | 0.38 |
ENSDART00000108579
|
blzf1
|
basic leucine zipper nuclear factor 1 |
| chr6_-_25065745 | 0.37 |
ENSDART00000165170
|
znf326
|
zinc finger protein 326 |
| chr17_+_15527861 | 0.37 |
ENSDART00000148443
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr19_+_7082199 | 0.35 |
ENSDART00000110366
|
zbtb22b
|
zinc finger and BTB domain containing 22b |
| chr7_+_25778794 | 0.35 |
ENSDART00000173611
|
si:dkey-6n21.12
|
si:dkey-6n21.12 |
| chr11_-_18637646 | 0.30 |
ENSDART00000156276
|
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr21_-_5731575 | 0.29 |
ENSDART00000172650
|
wu:fj64h06
|
wu:fj64h06 |
| chr15_-_20189097 | 0.28 |
ENSDART00000152355
|
med13b
|
mediator complex subunit 13b |
| chr16_-_47366405 | 0.27 |
ENSDART00000169697
|
mios
|
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr3_+_13780104 | 0.27 |
ENSDART00000164179
|
syce2
|
synaptonemal complex central element protein 2 |
| chr19_-_42847112 | 0.26 |
ENSDART00000086961
|
vps45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr7_+_61938347 | 0.25 |
ENSDART00000025308
|
tbc1d19
|
TBC1 domain family, member 19 |
| chr18_-_3455306 | 0.25 |
ENSDART00000166841
ENSDART00000161197 |
dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr13_+_25394 | 0.25 |
ENSDART00000102511
ENSDART00000159546 |
cfap36
|
cilia and flagella associated protein 36 |
| chr13_-_10488076 | 0.23 |
ENSDART00000160561
|
si:ch73-54n14.2
|
si:ch73-54n14.2 |
| chr17_+_26785230 | 0.23 |
ENSDART00000023470
|
pgrmc2
|
progesterone receptor membrane component 2 |
| chr7_+_39393052 | 0.22 |
ENSDART00000146171
|
zgc:158564
|
zgc:158564 |
| chr8_-_22252628 | 0.22 |
ENSDART00000121513
|
nphp4
|
nephronophthisis 4 |
| chr15_-_20189405 | 0.21 |
ENSDART00000152355
|
med13b
|
mediator complex subunit 13b |
| chr1_+_22583970 | 0.21 |
ENSDART00000036448
|
lias
|
lipoic acid synthetase |
| chr5_+_31541677 | 0.21 |
ENSDART00000041504
|
tescb
|
tescalcin b |
| chr8_-_11039536 | 0.20 |
ENSDART00000143844
|
dennd2c
|
DENN/MADD domain containing 2C |
| chr1_+_499101 | 0.20 |
ENSDART00000108579
|
blzf1
|
basic leucine zipper nuclear factor 1 |
| chr6_-_53391354 | 0.19 |
ENSDART00000155483
|
si:ch211-161c3.6
|
si:ch211-161c3.6 |
| chr23_+_41935781 | 0.19 |
ENSDART00000171885
|
scp2b
|
sterol carrier protein 2b |
| chr13_+_30939809 | 0.19 |
ENSDART00000110560
|
si:ch211-223a10.1
|
si:ch211-223a10.1 |
| chr6_-_25065773 | 0.18 |
ENSDART00000165170
|
znf326
|
zinc finger protein 326 |
| chr18_-_35861482 | 0.17 |
ENSDART00000088488
|
opa3
|
optic atrophy 3 |
| chr7_-_54752472 | 0.17 |
ENSDART00000172394
ENSDART00000170637 |
tpcn2
|
two pore segment channel 2 |
| chr7_-_54752678 | 0.17 |
ENSDART00000172394
ENSDART00000170637 |
tpcn2
|
two pore segment channel 2 |
| chr2_+_47885621 | 0.17 |
|
|
|
| chr10_-_16069787 | 0.16 |
ENSDART00000122540
|
aldh7a1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr16_-_38407365 | 0.16 |
|
|
|
| chr8_-_11039421 | 0.16 |
ENSDART00000143844
|
dennd2c
|
DENN/MADD domain containing 2C |
| chr6_-_54340370 | 0.15 |
ENSDART00000156501
|
zgc:101577
|
zgc:101577 |
| chr21_+_1084935 | 0.14 |
|
|
|
| chr7_-_54752565 | 0.12 |
ENSDART00000172394
ENSDART00000170637 |
tpcn2
|
two pore segment channel 2 |
| chr10_-_8476067 | 0.10 |
ENSDART00000108643
|
tctn1
|
tectonic family member 1 |
| chr15_+_1801642 | 0.09 |
ENSDART00000168250
|
cul3b
|
cullin 3b |
| chr13_+_9036533 | 0.09 |
ENSDART00000109126
|
alms1
|
Alstrom syndrome protein 1 |
| chr17_-_24420654 | 0.08 |
|
|
|
| chr4_+_13651618 | 0.08 |
ENSDART00000110370
|
nrcama
|
neuronal cell adhesion molecule a |
| chr10_+_36148811 | 0.07 |
ENSDART00000086490
|
smyd4
|
SET and MYND domain containing 4 |
| chr3_+_13780013 | 0.05 |
ENSDART00000164179
|
syce2
|
synaptonemal complex central element protein 2 |
| chr7_+_29592486 | 0.05 |
ENSDART00000134068
|
tln2a
|
talin 2a |
| chr1_+_33471274 | 0.04 |
ENSDART00000046094
|
arl6
|
ADP-ribosylation factor-like 6 |
| chr20_+_36779159 | 0.03 |
ENSDART00000130513
|
ncoa1
|
nuclear receptor coactivator 1 |
| chr22_-_121198 | 0.03 |
|
|
|
| chr2_+_16505452 | 0.02 |
ENSDART00000045933
|
sh3glb1b
|
SH3-domain GRB2-like endophilin B1b |
| chr8_-_36585594 | 0.02 |
ENSDART00000047912
ENSDART00000127435 |
gpkow
|
G patch domain and KOW motifs |
| chr7_+_29592666 | 0.01 |
ENSDART00000134068
|
tln2a
|
talin 2a |
| chr19_-_42847182 | 0.01 |
ENSDART00000086961
|
vps45
|
vacuolar protein sorting 45 homolog (S. cerevisiae) |
| chr7_+_29592358 | 0.01 |
ENSDART00000167286
|
tln2a
|
talin 2a |
| chr1_-_499082 | 0.00 |
ENSDART00000144406
ENSDART00000031635 |
ercc5
|
excision repair cross-complementation group 5 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0071932 | replication fork reversal(GO:0071932) |
| 0.3 | 0.9 | GO:2001284 | epithelial cell morphogenesis involved in gastrulation(GO:0003381) BMP secretion(GO:0038055) positive regulation of BMP secretion(GO:1900144) regulation of BMP secretion(GO:2001284) |
| 0.1 | 0.4 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.1 | 0.5 | GO:2000290 | regulation of myotome development(GO:2000290) |
| 0.1 | 0.3 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.1 | 0.5 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.1 | 0.2 | GO:0030728 | ovulation(GO:0030728) |
| 0.1 | 0.8 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.3 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.2 | GO:0032367 | intracellular cholesterol transport(GO:0032367) positive regulation of lipid transport(GO:0032370) |
| 0.0 | 0.3 | GO:0007130 | synaptonemal complex assembly(GO:0007130) synaptonemal complex organization(GO:0070193) |
| 0.0 | 0.2 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.6 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 1.0 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.3 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.1 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.5 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0005760 | gamma DNA polymerase complex(GO:0005760) |
| 0.3 | 1.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.1 | 0.3 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.4 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.9 | GO:0032432 | actin filament bundle(GO:0032432) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0016890 | site-specific endodeoxyribonuclease activity, specific for altered base(GO:0016890) |
| 0.1 | 0.4 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.5 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.8 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 1.0 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.3 | GO:0001047 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.0 | 0.5 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |