Project
DANIO-CODE
Navigation
Downloads
Results for rfx7a+rfx7b
Z-value: 0.56
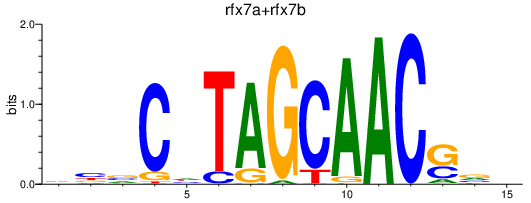
Transcription factors associated with rfx7a+rfx7b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rfx7a
|
ENSDARG00000077237 | regulatory factor X7a |
|
rfx7b
|
ENSDARG00000103691 | regulatory factor X7b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ENSDARG00000103691 | dr10_dc_chr25_+_365026_365133 | -0.66 | 5.0e-03 | Click! |
| rfx7 | dr10_dc_chr7_+_34235330_34235412 | -0.65 | 6.6e-03 | Click! |
Activity profile of rfx7a+rfx7b motif
Sorted Z-values of rfx7a+rfx7b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of rfx7a+rfx7b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_36721626 | 2.44 |
ENSDART00000017188
ENSDART00000124698 |
ncl
|
nucleolin |
| chr24_-_9857510 | 1.25 |
ENSDART00000136274
|
si:ch211-146l10.7
|
si:ch211-146l10.7 |
| chr24_-_9865837 | 1.16 |
ENSDART00000106244
|
zgc:171750
|
zgc:171750 |
| chr14_-_35552296 | 1.13 |
ENSDART00000052648
|
tmem144b
|
transmembrane protein 144b |
| chr14_+_24543399 | 1.06 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr19_-_28004606 | 0.93 |
ENSDART00000121643
|
si:dkeyp-46h3.2
|
si:dkeyp-46h3.2 |
| chr2_-_45810787 | 0.92 |
ENSDART00000135665
|
prpf38b
|
pre-mRNA processing factor 38B |
| chr10_+_44845652 | 0.85 |
ENSDART00000157458
|
ubc
|
ubiquitin C |
| chr17_-_2544896 | 0.76 |
ENSDART00000109582
|
spata7
|
spermatogenesis associated 7 |
| chr7_+_40866623 | 0.71 |
ENSDART00000052274
|
puf60b
|
poly-U binding splicing factor b |
| chr7_+_51520610 | 0.70 |
ENSDART00000174201
|
slc38a7
|
solute carrier family 38, member 7 |
| chr1_-_51075777 | 0.68 |
ENSDART00000152745
|
snrpb2
|
small nuclear ribonucleoprotein polypeptide B2 |
| chr8_-_14042703 | 0.64 |
ENSDART00000042867
|
dedd
|
death effector domain containing |
| chr25_+_34340569 | 0.64 |
ENSDART00000157638
|
tmem231
|
transmembrane protein 231 |
| chr19_+_14489756 | 0.62 |
ENSDART00000164386
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr6_+_21887935 | 0.61 |
ENSDART00000157796
|
cbx8b
|
chromobox homolog 8b |
| chr13_-_6195363 | 0.59 |
ENSDART00000148907
|
mcph1
|
microcephalin 1 |
| chr8_+_5222023 | 0.58 |
ENSDART00000035676
|
bnip3la
|
BCL2/adenovirus E1B interacting protein 3-like a |
| chr17_-_33764220 | 0.58 |
ENSDART00000043651
|
dnal1
|
dynein, axonemal, light chain 1 |
| chr4_-_14208573 | 0.57 |
ENSDART00000015134
|
twf1b
|
twinfilin actin-binding protein 1b |
| chr5_-_66145078 | 0.55 |
ENSDART00000041441
|
stip1
|
stress-induced phosphoprotein 1 |
| chr10_-_25725495 | 0.55 |
ENSDART00000177220
|
AL837524.2
|
ENSDARG00000108650 |
| chr3_+_1398333 | 0.54 |
ENSDART00000031823
|
triobpb
|
TRIO and F-actin binding protein b |
| chr13_-_36719000 | 0.54 |
ENSDART00000129562
ENSDART00000150899 |
nin
|
ninein (GSK3B interacting protein) |
| chr7_-_19989384 | 0.53 |
ENSDART00000173619
|
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr8_-_16479425 | 0.51 |
ENSDART00000146469
ENSDART00000132681 |
ttc39a
|
tetratricopeptide repeat domain 39A |
| chr14_-_33141111 | 0.51 |
ENSDART00000147059
|
lamp2
|
lysosomal-associated membrane protein 2 |
| chr12_+_46281623 | 0.51 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr12_+_46281592 | 0.51 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr7_-_19989419 | 0.50 |
ENSDART00000127699
|
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr22_-_36721592 | 0.50 |
ENSDART00000130171
|
ncl
|
nucleolin |
| chr23_-_20050419 | 0.48 |
ENSDART00000054659
|
fam58a
|
family with sequence similarity 58, member A |
| chr21_-_32748287 | 0.48 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr2_-_45810868 | 0.48 |
ENSDART00000135665
ENSDART00000075080 |
prpf38b
|
pre-mRNA processing factor 38B |
| chr16_+_54833107 | 0.47 |
ENSDART00000157641
|
fbxo43
|
F-box protein 43 |
| chr19_-_5142209 | 0.47 |
ENSDART00000150980
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr7_+_17656192 | 0.47 |
ENSDART00000077113
|
mta2
|
metastasis associated 1 family, member 2 |
| chr5_+_29791993 | 0.46 |
ENSDART00000086765
|
stk36
|
serine/threonine kinase 36 (fused homolog, Drosophila) |
| chr16_+_25693740 | 0.45 |
ENSDART00000077484
|
zhx2a
|
zinc fingers and homeoboxes 2a |
| chr19_-_27811300 | 0.45 |
ENSDART00000103940
|
mgat1b
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase b |
| chr13_-_48473254 | 0.44 |
ENSDART00000167157
|
si:ch1073-266p11.2
|
si:ch1073-266p11.2 |
| chr11_+_37385705 | 0.44 |
ENSDART00000129918
|
kif17
|
kinesin family member 17 |
| chr12_+_46281511 | 0.44 |
ENSDART00000149326
|
ush1gb
|
Usher syndrome 1Gb (autosomal recessive) |
| chr13_-_24586850 | 0.44 |
ENSDART00000172156
|
erlin1
|
ER lipid raft associated 1 |
| chr3_+_39413882 | 0.44 |
ENSDART00000156848
|
BX537342.1
|
ENSDARG00000097765 |
| chr6_+_4000356 | 0.43 |
ENSDART00000111817
|
trim25l
|
tripartite motif containing 25, like |
| chr10_+_25234636 | 0.43 |
ENSDART00000024514
|
chordc1b
|
cysteine and histidine-rich domain (CHORD) containing 1b |
| chr17_+_23948060 | 0.42 |
|
|
|
| chr9_+_55126932 | 0.42 |
ENSDART00000164812
|
ofd1
|
oral-facial-digital syndrome 1 |
| chr17_-_2544927 | 0.41 |
ENSDART00000109582
|
spata7
|
spermatogenesis associated 7 |
| chr14_-_32755189 | 0.40 |
ENSDART00000074720
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr3_-_31487744 | 0.40 |
ENSDART00000124559
|
moto
|
minamoto |
| chr3_+_1398365 | 0.40 |
ENSDART00000031823
|
triobpb
|
TRIO and F-actin binding protein b |
| chr8_+_45285797 | 0.39 |
ENSDART00000136234
|
ubap2b
|
ubiquitin associated protein 2b |
| chr25_+_27300835 | 0.38 |
ENSDART00000103519
|
wasla
|
Wiskott-Aldrich syndrome-like a |
| chr11_-_40255430 | 0.37 |
ENSDART00000127932
|
miip
|
migration and invasion inhibitory protein |
| chr22_-_11817641 | 0.37 |
ENSDART00000000192
|
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr12_+_10668405 | 0.37 |
ENSDART00000161986
ENSDART00000158227 |
top2a
|
topoisomerase (DNA) II alpha |
| chr14_-_30564987 | 0.37 |
ENSDART00000173451
|
si:ch211-126c2.4
|
si:ch211-126c2.4 |
| chr21_+_21754830 | 0.37 |
ENSDART00000151654
|
neu3.1
|
sialidase 3 (membrane sialidase), tandem duplicate 1 |
| chr9_-_26776407 | 0.36 |
|
|
|
| chr5_-_24378527 | 0.36 |
ENSDART00000057248
|
ewsr1b
|
EWS RNA-binding protein 1b |
| chr21_+_26689954 | 0.36 |
ENSDART00000065392
|
calm3b
|
calmodulin 3b (phosphorylase kinase, delta) |
| chr13_+_24467302 | 0.36 |
ENSDART00000158949
|
AL831745.3
|
ENSDARG00000101436 |
| chr22_-_11817587 | 0.36 |
ENSDART00000000192
|
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr7_+_17656099 | 0.36 |
ENSDART00000077113
|
mta2
|
metastasis associated 1 family, member 2 |
| chr23_-_27768575 | 0.36 |
ENSDART00000146703
|
IKZF4
|
IKAROS family zinc finger 4 |
| chr5_+_36049836 | 0.36 |
ENSDART00000147667
|
alkbh6
|
alkB homolog 6 |
| chr18_-_20741085 | 0.35 |
|
|
|
| chr15_-_37687982 | 0.35 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr6_-_7854902 | 0.34 |
|
|
|
| chr15_+_19948916 | 0.33 |
ENSDART00000054416
|
dyrk1ab
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, b |
| chr1_-_52642562 | 0.33 |
ENSDART00000150579
|
znf330
|
zinc finger protein 330 |
| chr19_-_6401104 | 0.33 |
|
|
|
| chr24_-_5904097 | 0.32 |
ENSDART00000007373
ENSDART00000135124 |
acbd5a
|
acyl-CoA binding domain containing 5a |
| chr8_+_5222145 | 0.32 |
ENSDART00000035676
|
bnip3la
|
BCL2/adenovirus E1B interacting protein 3-like a |
| chr18_-_44365869 | 0.32 |
ENSDART00000166935
|
prdm10
|
PR domain containing 10 |
| chr5_+_36049885 | 0.31 |
ENSDART00000147667
|
alkbh6
|
alkB homolog 6 |
| chr11_-_40255517 | 0.31 |
ENSDART00000127932
|
miip
|
migration and invasion inhibitory protein |
| chr14_-_38487071 | 0.31 |
ENSDART00000160000
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr20_+_49332270 | 0.31 |
|
|
|
| chr14_+_35552581 | 0.30 |
ENSDART00000135079
|
zgc:63568
|
zgc:63568 |
| chr19_-_5142238 | 0.30 |
ENSDART00000130062
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr4_-_72771281 | 0.30 |
ENSDART00000164550
ENSDART00000174320 |
RAB21
|
RAB21, member RAS oncogene family |
| chr23_+_7776647 | 0.30 |
ENSDART00000161193
|
kif3b
|
kinesin family member 3B |
| chr8_+_45285975 | 0.30 |
ENSDART00000137104
|
ubap2b
|
ubiquitin associated protein 2b |
| chr15_+_19948869 | 0.29 |
ENSDART00000054416
|
dyrk1ab
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, b |
| chr25_-_13562558 | 0.29 |
ENSDART00000045488
|
csnk2a2b
|
casein kinase 2, alpha prime polypeptide b |
| chr24_+_14095601 | 0.29 |
ENSDART00000124740
|
ncoa2
|
nuclear receptor coactivator 2 |
| chr7_+_32204099 | 0.28 |
ENSDART00000173966
|
BX511109.2
|
ENSDARG00000105604 |
| chr3_+_29338315 | 0.28 |
ENSDART00000103592
|
fam83fa
|
family with sequence similarity 83, member Fa |
| KN149789v1_-_7491 | 0.28 |
|
|
|
| chr19_-_27811272 | 0.28 |
ENSDART00000103940
|
mgat1b
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase b |
| chr11_+_1558144 | 0.28 |
ENSDART00000154583
|
si:dkey-40c23.2
|
si:dkey-40c23.2 |
| chr19_+_10742887 | 0.28 |
ENSDART00000165653
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr22_-_24957034 | 0.27 |
ENSDART00000142147
|
dnal4b
|
dynein, axonemal, light chain 4b |
| chr1_-_52642446 | 0.27 |
ENSDART00000131520
|
znf330
|
zinc finger protein 330 |
| chr12_+_1154951 | 0.27 |
ENSDART00000160868
|
ENSDARG00000102861
|
ENSDARG00000102861 |
| chr1_-_52642622 | 0.27 |
ENSDART00000150579
|
znf330
|
zinc finger protein 330 |
| chr2_+_16780992 | 0.25 |
ENSDART00000005381
|
zgc:110269
|
zgc:110269 |
| chr17_-_45395144 | 0.25 |
ENSDART00000132969
|
znf106a
|
zinc finger protein 106a |
| chr18_-_44366105 | 0.25 |
ENSDART00000166935
|
prdm10
|
PR domain containing 10 |
| chr16_+_26727466 | 0.24 |
ENSDART00000087537
|
epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chr18_-_44366076 | 0.24 |
ENSDART00000166935
|
prdm10
|
PR domain containing 10 |
| chr1_+_9621612 | 0.24 |
ENSDART00000043881
|
zgc:77880
|
zgc:77880 |
| chr15_-_37687790 | 0.24 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr20_-_44678938 | 0.24 |
ENSDART00000148639
|
ubxn2a
|
UBX domain protein 2A |
| chr21_+_11943637 | 0.24 |
ENSDART00000141306
|
zgc:162344
|
zgc:162344 |
| chr11_-_26352615 | 0.24 |
ENSDART00000154349
ENSDART00000002846 |
st3gal8
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 8 |
| chr11_+_1506660 | 0.24 |
ENSDART00000066191
|
ift52
|
intraflagellar transport 52 homolog (Chlamydomonas) |
| chr16_+_10950134 | 0.23 |
ENSDART00000065467
|
dedd1
|
death effector domain-containing 1 |
| chr13_+_24549249 | 0.23 |
ENSDART00000113981
|
zgc:66426
|
zgc:66426 |
| chr4_-_3215313 | 0.23 |
ENSDART00000112210
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr19_+_10742387 | 0.23 |
ENSDART00000091813
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr17_+_24666346 | 0.23 |
ENSDART00000146309
|
znf593
|
zinc finger protein 593 |
| chr13_+_25394 | 0.23 |
ENSDART00000102511
ENSDART00000159546 |
cfap36
|
cilia and flagella associated protein 36 |
| chr5_-_3323004 | 0.23 |
|
|
|
| chr11_+_25090547 | 0.22 |
ENSDART00000141478
|
fam83d
|
family with sequence similarity 83, member D |
| chr15_+_22331719 | 0.22 |
ENSDART00000110665
|
nrgnb
|
neurogranin (protein kinase C substrate, RC3) b |
| chr19_+_10742807 | 0.22 |
ENSDART00000165653
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr8_-_22492972 | 0.22 |
|
|
|
| chr14_+_23412042 | 0.22 |
ENSDART00000054266
|
gnpda1
|
glucosamine-6-phosphate deaminase 1 |
| chr7_-_39467720 | 0.21 |
ENSDART00000052201
|
ccdc96
|
coiled-coil domain containing 96 |
| chr11_+_1506720 | 0.21 |
ENSDART00000066191
|
ift52
|
intraflagellar transport 52 homolog (Chlamydomonas) |
| chr4_-_72771112 | 0.21 |
ENSDART00000164550
ENSDART00000174320 |
RAB21
|
RAB21, member RAS oncogene family |
| chr24_+_12845339 | 0.21 |
ENSDART00000126842
|
flj11011l
|
hypothetical protein FLJ11011-like (H. sapiens) |
| chr17_-_33764256 | 0.21 |
ENSDART00000043651
|
dnal1
|
dynein, axonemal, light chain 1 |
| chr23_+_24575330 | 0.20 |
ENSDART00000078824
|
szrd1
|
SUZ RNA binding domain containing 1 |
| chr4_+_90885 | 0.20 |
ENSDART00000169805
|
eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr12_+_19066653 | 0.20 |
ENSDART00000134726
|
cby1
|
chibby homolog 1 (Drosophila) |
| chr8_+_45286071 | 0.20 |
ENSDART00000137104
|
ubap2b
|
ubiquitin associated protein 2b |
| chr13_+_9036533 | 0.20 |
ENSDART00000109126
|
alms1
|
Alstrom syndrome protein 1 |
| chr17_-_43437795 | 0.20 |
ENSDART00000043866
|
plk4
|
polo-like kinase 4 (Drosophila) |
| chr1_-_52642475 | 0.19 |
ENSDART00000131520
|
znf330
|
zinc finger protein 330 |
| chr1_+_9621488 | 0.19 |
ENSDART00000043881
|
zgc:77880
|
zgc:77880 |
| chr14_-_25636795 | 0.19 |
|
|
|
| chr8_-_22252628 | 0.19 |
ENSDART00000121513
|
nphp4
|
nephronophthisis 4 |
| chr17_-_28794619 | 0.19 |
ENSDART00000001444
|
g2e3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr23_-_29431197 | 0.19 |
ENSDART00000156512
|
si:ch211-129o18.4
|
si:ch211-129o18.4 |
| chr24_-_15313198 | 0.19 |
|
|
|
| chr24_-_5903682 | 0.19 |
ENSDART00000007373
ENSDART00000135124 |
acbd5a
|
acyl-CoA binding domain containing 5a |
| chr3_+_45375580 | 0.19 |
ENSDART00000162799
|
crb3a
|
crumbs homolog 3a |
| chr9_+_23118780 | 0.18 |
ENSDART00000061299
|
tsn
|
translin |
| chr12_-_28840748 | 0.18 |
|
|
|
| chr3_+_21059313 | 0.18 |
ENSDART00000141410
|
zgc:123295
|
zgc:123295 |
| chr19_+_9066225 | 0.18 |
ENSDART00000018973
|
scamp3
|
secretory carrier membrane protein 3 |
| chr21_+_11943609 | 0.17 |
ENSDART00000155426
ENSDART00000102463 |
zgc:162344
|
zgc:162344 |
| chr16_+_10950160 | 0.17 |
ENSDART00000065467
|
dedd1
|
death effector domain-containing 1 |
| chr11_-_40255633 | 0.17 |
ENSDART00000172819
|
miip
|
migration and invasion inhibitory protein |
| chr21_+_10152870 | 0.16 |
|
|
|
| chr14_+_1342695 | 0.16 |
|
|
|
| chr19_+_10742594 | 0.16 |
ENSDART00000091813
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr7_+_17655778 | 0.15 |
ENSDART00000077113
|
mta2
|
metastasis associated 1 family, member 2 |
| chr1_+_5527701 | 0.15 |
ENSDART00000145378
|
fastkd2
|
FAST kinase domains 2 |
| chr19_+_10742344 | 0.15 |
ENSDART00000091813
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr4_-_1647837 | 0.15 |
ENSDART00000178560
|
ENSDARG00000063672
|
ENSDARG00000063672 |
| chr15_+_19948426 | 0.15 |
ENSDART00000054416
|
dyrk1ab
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A, b |
| chr5_-_57053661 | 0.14 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr25_-_13562597 | 0.14 |
ENSDART00000045488
|
csnk2a2b
|
casein kinase 2, alpha prime polypeptide b |
| chr3_+_52745606 | 0.14 |
ENSDART00000104683
|
pbx4
|
pre-B-cell leukemia transcription factor 4 |
| chr6_+_21887963 | 0.14 |
ENSDART00000157796
|
cbx8b
|
chromobox homolog 8b |
| chr17_-_28794581 | 0.13 |
ENSDART00000132811
|
g2e3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr15_-_37687938 | 0.13 |
ENSDART00000154641
|
proser3
|
proline and serine rich 3 |
| chr16_+_28643677 | 0.13 |
ENSDART00000149306
|
nmt2
|
N-myristoyltransferase 2 |
| chr6_-_31752797 | 0.13 |
ENSDART00000087964
|
cachd1
|
cache domain containing 1 |
| chr8_+_31006910 | 0.13 |
ENSDART00000124213
|
odf2b
|
outer dense fiber of sperm tails 2b |
| chr7_+_51883949 | 0.13 |
ENSDART00000130282
|
katnb1
|
katanin p80 (WD repeat containing) subunit B 1 |
| chr19_+_5562075 | 0.13 |
ENSDART00000148794
|
jupb
|
junction plakoglobin b |
| chr7_+_40866459 | 0.13 |
ENSDART00000052274
|
puf60b
|
poly-U binding splicing factor b |
| chr23_+_29431388 | 0.13 |
ENSDART00000027255
|
tardbpl
|
TAR DNA binding protein, like |
| chr24_+_36351542 | 0.12 |
ENSDART00000062745
|
riok3
|
RIO kinase 3 (yeast) |
| chr18_-_21961658 | 0.12 |
ENSDART00000043452
|
tsnaxip1
|
translin-associated factor X interacting protein 1 |
| chr2_-_16780900 | 0.12 |
ENSDART00000144801
|
atr
|
ATR serine/threonine kinase |
| chr9_-_41151588 | 0.12 |
ENSDART00000135805
|
BX323586.2
|
ENSDARG00000092912 |
| chr11_+_1506452 | 0.12 |
ENSDART00000066191
|
ift52
|
intraflagellar transport 52 homolog (Chlamydomonas) |
| chr25_-_10532989 | 0.12 |
ENSDART00000177834
|
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr22_-_20695416 | 0.12 |
ENSDART00000105532
|
oaz1a
|
ornithine decarboxylase antizyme 1a |
| chr7_-_40467922 | 0.11 |
ENSDART00000084179
|
rnf32
|
ring finger protein 32 |
| chr25_-_17282337 | 0.11 |
ENSDART00000064586
|
cyp2x7
|
cytochrome P450, family 2, subfamily X, polypeptide 7 |
| chr17_+_33344090 | 0.11 |
ENSDART00000123114
|
dnajc27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr6_+_6645278 | 0.11 |
ENSDART00000138283
|
dtd1
|
D-tyrosyl-tRNA deacylase 1 |
| chr13_-_33115645 | 0.11 |
ENSDART00000158580
|
rtf1
|
RTF1 homolog, Paf1/RNA polymerase II complex component |
| chr11_-_40154790 | 0.11 |
ENSDART00000086296
|
trim62
|
tripartite motif containing 62 |
| chr5_-_51351827 | 0.10 |
ENSDART00000097194
|
serinc5
|
serine incorporator 5 |
| chr7_-_19747123 | 0.10 |
ENSDART00000100798
|
trip6
|
thyroid hormone receptor interactor 6 |
| chr20_+_2113465 | 0.10 |
|
|
|
| chr16_+_24047649 | 0.10 |
|
|
|
| chr3_-_37445694 | 0.10 |
|
|
|
| chr20_+_14073428 | 0.10 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr3_-_37445791 | 0.09 |
|
|
|
| chr9_-_23346038 | 0.09 |
ENSDART00000135461
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr20_+_47639141 | 0.09 |
ENSDART00000043938
|
tram2
|
translocation associated membrane protein 2 |
| chr24_+_28571936 | 0.09 |
ENSDART00000153933
|
CR933730.1
|
ENSDARG00000097841 |
| chr25_+_34340352 | 0.09 |
ENSDART00000163190
|
tmem231
|
transmembrane protein 231 |
| chr14_-_21363222 | 0.09 |
ENSDART00000089845
ENSDART00000161713 |
kdm3b
|
lysine (K)-specific demethylase 3B |
| chr24_-_24859135 | 0.08 |
ENSDART00000136860
|
zdhhc20b
|
zinc finger, DHHC-type containing 20b |
| chr7_+_20046760 | 0.08 |
ENSDART00000131019
|
acadvl
|
acyl-CoA dehydrogenase, very long chain |
| chr9_-_7013144 | 0.08 |
ENSDART00000081718
|
slc9a2
|
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chr8_-_8307383 | 0.08 |
|
|
|
| chr3_-_37445761 | 0.08 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.3 | 2.9 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.2 | 0.9 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 1.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.5 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 1.4 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.1 | 0.6 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.1 | 0.4 | GO:0099640 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) axo-dendritic protein transport(GO:0099640) |
| 0.1 | 0.4 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.5 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) negative regulation of meiotic cell cycle(GO:0051447) |
| 0.1 | 0.5 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 1.0 | GO:0008625 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) |
| 0.1 | 0.4 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.2 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.1 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.1 | 0.4 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.1 | 1.0 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.1 | 0.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.5 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.5 | GO:0035794 | positive regulation of mitochondrial membrane permeability(GO:0035794) mitochondrial outer membrane permeabilization(GO:0097345) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.0 | 0.8 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.4 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.2 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.0 | 0.1 | GO:0018377 | N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.0 | 0.4 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.8 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.6 | GO:0031100 | organ regeneration(GO:0031100) |
| 0.0 | 0.2 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.0 | 0.3 | GO:0043584 | nose development(GO:0043584) |
| 0.0 | 0.2 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.0 | 0.4 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 1.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.4 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.1 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.6 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.5 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling(GO:1901224) |
| 0.0 | 0.2 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 1.0 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.1 | GO:0097369 | hypotonic response(GO:0006971) response to salt stress(GO:0009651) hypotonic salinity response(GO:0042539) sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0042073 | intraciliary transport(GO:0042073) protein transport along microtubule(GO:0098840) |
| 0.0 | 0.6 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 1.0 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.2 | 0.5 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.1 | 0.4 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 1.0 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 1.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 0.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.1 | 0.6 | GO:0035060 | brahma complex(GO:0035060) |
| 0.1 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 2.3 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.2 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 1.4 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.7 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.4 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.5 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.5 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 5.3 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.7 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.2 | 0.7 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.2 | 1.0 | GO:0090624 | endoribonuclease activity, cleaving miRNA-paired mRNA(GO:0090624) |
| 0.1 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.3 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.7 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 0.4 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.4 | GO:0052795 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.1 | 0.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 1.1 | GO:1901476 | carbohydrate transmembrane transporter activity(GO:0015144) carbohydrate transporter activity(GO:1901476) |
| 0.1 | 1.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.9 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.1 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.1 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.1 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.0 | 0.1 | GO:0005451 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.4 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0051219 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.8 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.0 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.5 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |