Project
DANIO-CODE
Navigation
Downloads
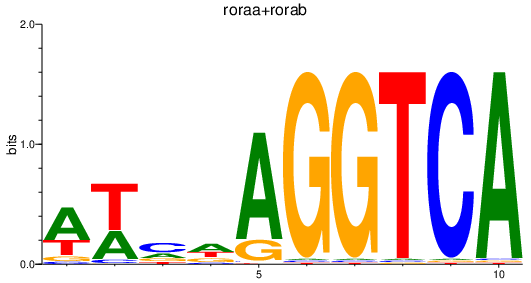
Results for roraa+rorab
Z-value: 0.39
Transcription factors associated with roraa+rorab
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rorab
|
ENSDARG00000001910 | RAR-related orphan receptor A, paralog b |
|
roraa
|
ENSDARG00000031768 | RAR-related orphan receptor A, paralog a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rorab | dr10_dc_chr7_-_29354323_29354384 | 0.44 | 8.8e-02 | Click! |
| roraa | dr10_dc_chr25_+_33364224_33364302 | 0.38 | 1.5e-01 | Click! |
Activity profile of roraa+rorab motif
Sorted Z-values of roraa+rorab motif
Network of associatons between targets according to the STRING database.
First level regulatory network of roraa+rorab
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_22881453 | 0.52 |
|
|
|
| chr25_-_30818836 | 0.50 |
ENSDART00000156828
|
prr33
|
proline rich 33 |
| chr6_-_39767452 | 0.48 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr21_+_7844259 | 0.45 |
ENSDART00000027268
|
otpa
|
orthopedia homeobox a |
| chr13_+_50062513 | 0.42 |
ENSDART00000124142
ENSDART00000099537 |
cox5b2
|
cytochrome c oxidase subunit Vb 2 |
| chr9_-_53220424 | 0.41 |
|
|
|
| chr11_+_30482530 | 0.39 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr23_+_20183765 | 0.38 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr15_-_12338925 | 0.36 |
ENSDART00000170093
|
si:dkey-36i7.3
|
si:dkey-36i7.3 |
| chr21_-_19789339 | 0.35 |
|
|
|
| chr2_+_42342148 | 0.35 |
ENSDART00000144716
|
cavin4a
|
caveolae associated protein 4a |
| chr1_-_50215233 | 0.34 |
ENSDART00000137648
|
si:dkeyp-123h10.2
|
si:dkeyp-123h10.2 |
| chr20_+_40247918 | 0.34 |
ENSDART00000121818
|
trdn
|
triadin |
| chr7_-_3900148 | 0.32 |
ENSDART00000019949
|
ndrg2
|
NDRG family member 2 |
| chr13_-_21557620 | 0.31 |
ENSDART00000063825
|
sprn
|
shadow of prion protein |
| chr18_+_36650892 | 0.31 |
ENSDART00000098980
|
znf296
|
zinc finger protein 296 |
| chr16_-_28921444 | 0.31 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr5_-_29818204 | 0.30 |
ENSDART00000098285
|
atf5a
|
activating transcription factor 5a |
| chr14_-_33114284 | 0.30 |
ENSDART00000109615
|
tmem255a
|
transmembrane protein 255A |
| chr12_+_30673985 | 0.29 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr12_-_43939028 | 0.28 |
ENSDART00000170723
|
zgc:112964
|
zgc:112964 |
| chr20_-_39002962 | 0.28 |
|
|
|
| chr1_+_30840735 | 0.28 |
ENSDART00000075286
|
slc2a15b
|
solute carrier family 2 (facilitated glucose transporter), member 15b |
| chr19_+_20392669 | 0.27 |
ENSDART00000052627
|
npvf
|
neuropeptide VF precursor |
| chr3_-_16056593 | 0.27 |
|
|
|
| chr19_-_5186692 | 0.27 |
ENSDART00000037007
|
tpi1a
|
triosephosphate isomerase 1a |
| chr22_-_10324166 | 0.27 |
ENSDART00000092050
|
stab1
|
stabilin 1 |
| chr10_+_37983342 | 0.27 |
ENSDART00000172548
|
bhlha9
|
basic helix-loop-helix family, member a9 |
| chr24_-_5995073 | 0.25 |
ENSDART00000040865
|
pdss1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr9_-_21257082 | 0.24 |
ENSDART00000124533
|
tbx15
|
T-box 15 |
| chr14_-_33114322 | 0.24 |
ENSDART00000173267
|
tmem255a
|
transmembrane protein 255A |
| chr7_+_39957385 | 0.24 |
ENSDART00000052222
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr24_-_21768269 | 0.24 |
ENSDART00000081178
|
c1qtnf9
|
C1q and TNF related 9 |
| chr4_+_14658179 | 0.23 |
ENSDART00000168152
|
abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr11_+_24874769 | 0.23 |
ENSDART00000147546
|
ndrg3a
|
ndrg family member 3a |
| chr10_+_20156511 | 0.22 |
ENSDART00000139722
|
dmtn
|
dematin actin binding protein |
| chr6_-_10086031 | 0.22 |
|
|
|
| chr4_-_17640519 | 0.21 |
ENSDART00000108814
|
nrip2
|
nuclear receptor interacting protein 2 |
| chr20_-_14784875 | 0.21 |
ENSDART00000063857
|
scrn2
|
secernin 2 |
| chr14_-_30150071 | 0.21 |
ENSDART00000173107
|
micu3b
|
mitochondrial calcium uptake family, member 3b |
| chr1_+_30840656 | 0.21 |
ENSDART00000075286
|
slc2a15b
|
solute carrier family 2 (facilitated glucose transporter), member 15b |
| chr11_+_39768864 | 0.21 |
ENSDART00000130278
|
si:dkey-264d12.1
|
si:dkey-264d12.1 |
| chr8_+_6533379 | 0.21 |
ENSDART00000138135
|
vsig8b
|
V-set and immunoglobulin domain containing 8b |
| chr7_-_29070193 | 0.21 |
ENSDART00000147251
|
trpm1a
|
transient receptor potential cation channel, subfamily M, member 1a |
| chr7_-_3900223 | 0.20 |
ENSDART00000019949
|
ndrg2
|
NDRG family member 2 |
| chr11_+_39768718 | 0.19 |
ENSDART00000130278
|
si:dkey-264d12.1
|
si:dkey-264d12.1 |
| chr19_+_38033219 | 0.19 |
ENSDART00000158960
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr21_+_5775383 | 0.19 |
ENSDART00000165065
|
uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr17_+_33388313 | 0.19 |
ENSDART00000077553
|
xdh
|
xanthine dehydrogenase |
| chr18_-_48498261 | 0.18 |
ENSDART00000146346
|
kcnj1a.6
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 6 |
| chr24_+_29902165 | 0.17 |
ENSDART00000168422
ENSDART00000100092 ENSDART00000113304 |
frrs1b
|
ferric-chelate reductase 1b |
| chr13_+_42276937 | 0.16 |
|
|
|
| chr18_-_11439124 | 0.16 |
|
|
|
| chr4_-_1346961 | 0.16 |
ENSDART00000164623
|
ptn
|
pleiotrophin |
| chr18_-_15641443 | 0.16 |
ENSDART00000127614
|
arntl2
|
aryl hydrocarbon receptor nuclear translocator-like 2 |
| chr1_-_44476084 | 0.15 |
ENSDART00000034549
|
zgc:111983
|
zgc:111983 |
| chr7_+_51499409 | 0.15 |
ENSDART00000007767
|
got2b
|
glutamic-oxaloacetic transaminase 2b, mitochondrial |
| chr6_-_15444945 | 0.14 |
|
|
|
| chr7_+_30698873 | 0.14 |
ENSDART00000155974
|
tjp1a
|
tight junction protein 1a |
| chr17_-_7661762 | 0.14 |
ENSDART00000135538
|
rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr10_+_4924008 | 0.14 |
ENSDART00000108595
|
slc46a2
|
solute carrier family 46 member 2 |
| chr14_-_29518685 | 0.13 |
ENSDART00000136380
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr22_+_18521411 | 0.13 |
|
|
|
| chr18_+_17564926 | 0.13 |
|
|
|
| chr6_-_39767422 | 0.13 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr11_+_30482504 | 0.12 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr3_+_13487523 | 0.12 |
ENSDART00000166000
|
si:ch211-194b1.1
|
si:ch211-194b1.1 |
| chr22_+_29694605 | 0.12 |
|
|
|
| chr25_-_21058459 | 0.11 |
ENSDART00000156257
|
wnk1a
|
WNK lysine deficient protein kinase 1a |
| chr7_+_28862183 | 0.11 |
ENSDART00000052346
|
gnao1b
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, b |
| chr7_-_23775835 | 0.11 |
ENSDART00000144616
|
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr3_+_44928592 | 0.11 |
ENSDART00000168864
|
zgc:112146
|
zgc:112146 |
| chr3_-_26060748 | 0.11 |
ENSDART00000144726
|
ypel3
|
yippee-like 3 |
| chr12_-_24711169 | 0.11 |
ENSDART00000066317
|
foxn2b
|
forkhead box N2b |
| chr19_+_21140010 | 0.11 |
ENSDART00000090757
|
kat2b
|
K(lysine) acetyltransferase 2B |
| chr11_+_24662538 | 0.11 |
ENSDART00000148023
|
timm17a
|
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr17_+_30431520 | 0.10 |
ENSDART00000153826
|
lpin1
|
lipin 1 |
| chr16_-_30775394 | 0.10 |
|
|
|
| chr18_-_12483329 | 0.10 |
ENSDART00000092522
|
si:ch211-1e14.1
|
si:ch211-1e14.1 |
| chr10_+_4924065 | 0.10 |
ENSDART00000108595
|
slc46a2
|
solute carrier family 46 member 2 |
| chr2_-_14757989 | 0.10 |
ENSDART00000162816
|
BX510915.1
|
ENSDARG00000099630 |
| chr3_+_1413640 | 0.10 |
ENSDART00000055430
|
ndufa6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 |
| chr14_-_4166292 | 0.09 |
ENSDART00000127318
|
frmpd1b
|
FERM and PDZ domain containing 1b |
| chr7_-_33080261 | 0.09 |
ENSDART00000114041
|
anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr7_-_29300402 | 0.09 |
ENSDART00000099477
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr18_-_11439216 | 0.09 |
|
|
|
| chr23_-_24562107 | 0.09 |
ENSDART00000155593
|
tmem82
|
transmembrane protein 82 |
| chr24_+_37839153 | 0.09 |
|
|
|
| chr23_+_42920817 | 0.09 |
ENSDART00000055577
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr15_+_5124690 | 0.09 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr6_-_42951571 | 0.09 |
ENSDART00000003298
|
edem1
|
ER degradation enhancer, mannosidase alpha-like 1 |
| chr18_+_45574464 | 0.09 |
|
|
|
| chr19_+_44073707 | 0.09 |
ENSDART00000051723
|
si:ch211-193k19.1
|
si:ch211-193k19.1 |
| chr6_+_9801649 | 0.08 |
ENSDART00000149537
|
sucla2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr13_+_15669924 | 0.08 |
ENSDART00000146234
|
apopt1
|
apoptogenic 1, mitochondrial |
| chr20_+_45531190 | 0.08 |
|
|
|
| chr8_+_48888672 | 0.08 |
|
|
|
| chr17_-_7661725 | 0.08 |
ENSDART00000143870
|
rmnd1
|
required for meiotic nuclear division 1 homolog |
| chr6_-_45993454 | 0.07 |
ENSDART00000154392
|
ca16b
|
carbonic anhydrase XVI b |
| chr17_-_45140696 | 0.07 |
|
|
|
| chr11_+_31346560 | 0.07 |
ENSDART00000124830
ENSDART00000162768 |
zgc:162816
|
zgc:162816 |
| chr7_-_50546498 | 0.07 |
|
|
|
| chr6_+_40525779 | 0.07 |
ENSDART00000033819
|
prkcda
|
protein kinase C, delta a |
| chr1_+_23093114 | 0.07 |
|
|
|
| chr7_+_48532096 | 0.07 |
|
|
|
| chr18_+_29984261 | 0.07 |
|
|
|
| chr5_-_68751287 | 0.07 |
ENSDART00000112692
|
ENSDARG00000077155
|
ENSDARG00000077155 |
| chr7_+_22521055 | 0.07 |
ENSDART00000146801
|
rbm4.3
|
RNA binding motif protein 4.3 |
| chr1_+_58608231 | 0.07 |
ENSDART00000161872
ENSDART00000160658 |
parn
|
poly(A)-specific ribonuclease (deadenylation nuclease) |
| chr6_-_40314512 | 0.07 |
ENSDART00000033844
|
col7a1
|
collagen, type VII, alpha 1 |
| chr14_+_23220869 | 0.07 |
ENSDART00000112930
|
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr5_-_54773058 | 0.06 |
ENSDART00000145791
|
prune2
|
prune homolog 2 (Drosophila) |
| chr14_-_14353451 | 0.06 |
ENSDART00000170355
ENSDART00000159888 |
nsdhl
|
NAD(P) dependent steroid dehydrogenase-like |
| chr1_+_46640008 | 0.06 |
ENSDART00000053284
|
bcl9
|
B-cell CLL/lymphoma 9 |
| chr8_-_7547623 | 0.06 |
ENSDART00000178825
|
hcfc1b
|
host cell factor C1b |
| chr11_+_24662673 | 0.06 |
ENSDART00000148023
|
timm17a
|
translocase of inner mitochondrial membrane 17 homolog A (yeast) |
| chr1_+_16834347 | 0.06 |
ENSDART00000005593
ENSDART00000140076 |
casp3a
|
caspase 3, apoptosis-related cysteine peptidase a |
| chr14_-_4117151 | 0.06 |
|
|
|
| chr7_-_29956782 | 0.06 |
ENSDART00000173514
|
znf710b
|
zinc finger protein 710b |
| chr8_+_7974156 | 0.06 |
|
|
|
| chr7_-_55879342 | 0.06 |
ENSDART00000098438
|
spg7
|
spastic paraplegia 7 |
| chr14_+_33542705 | 0.06 |
ENSDART00000019396
|
clic2
|
chloride intracellular channel 2 |
| chr7_-_48532099 | 0.05 |
ENSDART00000015884
|
mfge8a
|
milk fat globule-EGF factor 8 protein a |
| chr21_+_37668284 | 0.05 |
ENSDART00000131188
|
nsd1b
|
nuclear receptor binding SET domain protein 1b |
| chr7_-_55879197 | 0.05 |
ENSDART00000098438
|
spg7
|
spastic paraplegia 7 |
| chr6_-_1562203 | 0.05 |
ENSDART00000067586
|
chchd6b
|
coiled-coil-helix-coiled-coil-helix domain containing 6b |
| chr6_-_10600358 | 0.05 |
ENSDART00000135065
|
atp5g3b
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit c3 (subunit 9) b |
| chr7_-_48532462 | 0.05 |
ENSDART00000015884
|
mfge8a
|
milk fat globule-EGF factor 8 protein a |
| chr7_+_26730804 | 0.05 |
|
|
|
| chr22_+_39067994 | 0.05 |
|
|
|
| chr20_-_53123015 | 0.05 |
ENSDART00000162812
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr25_-_21058792 | 0.05 |
ENSDART00000156257
|
wnk1a
|
WNK lysine deficient protein kinase 1a |
| chr3_-_26060787 | 0.05 |
ENSDART00000113843
|
ypel3
|
yippee-like 3 |
| chr14_-_15776937 | 0.05 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr18_-_15641870 | 0.04 |
ENSDART00000127614
|
arntl2
|
aryl hydrocarbon receptor nuclear translocator-like 2 |
| chr19_-_5416317 | 0.04 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr7_+_13238684 | 0.04 |
ENSDART00000053535
|
arih1l
|
ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 like |
| chr6_-_42951769 | 0.04 |
ENSDART00000147208
|
edem1
|
ER degradation enhancer, mannosidase alpha-like 1 |
| chr7_+_48532543 | 0.04 |
ENSDART00000145375
|
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr23_+_41935781 | 0.03 |
ENSDART00000171885
|
scp2b
|
sterol carrier protein 2b |
| chr10_-_4924455 | 0.03 |
|
|
|
| chr6_-_52484566 | 0.03 |
ENSDART00000112146
|
fam83c
|
family with sequence similarity 83, member C |
| chr7_+_39812659 | 0.03 |
ENSDART00000099046
|
ENSDARG00000036489
|
ENSDARG00000036489 |
| chr2_-_45118469 | 0.03 |
ENSDART00000018818
|
mul1a
|
mitochondrial E3 ubiquitin protein ligase 1a |
| chr18_+_9413668 | 0.03 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr6_-_39655998 | 0.03 |
ENSDART00000155859
|
larp4ab
|
La ribonucleoprotein domain family, member 4Ab |
| chr22_-_4867420 | 0.03 |
ENSDART00000132942
|
ncl1
|
nicalin |
| chr4_-_3340275 | 0.03 |
ENSDART00000009076
|
pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr2_+_10858655 | 0.03 |
ENSDART00000091570
|
fam69aa
|
family with sequence similarity 69, member Aa |
| chr24_-_8637129 | 0.02 |
ENSDART00000082362
|
mak
|
male germ cell-associated kinase |
| chr3_+_44928323 | 0.02 |
ENSDART00000170913
|
zgc:112146
|
zgc:112146 |
| chr8_+_21192955 | 0.02 |
ENSDART00000129210
|
cry1ba
|
cryptochrome circadian clock 1ba |
| chr4_-_9780352 | 0.02 |
ENSDART00000080744
|
svopl
|
SVOP-like |
| chr13_-_36719000 | 0.02 |
ENSDART00000129562
ENSDART00000150899 |
nin
|
ninein (GSK3B interacting protein) |
| chr12_-_24711074 | 0.02 |
ENSDART00000066317
|
foxn2b
|
forkhead box N2b |
| chr12_+_18407435 | 0.02 |
ENSDART00000146260
|
msrb1b
|
methionine sulfoxide reductase B1b |
| chr12_+_23742199 | 0.02 |
|
|
|
| chr8_-_49227232 | 0.02 |
|
|
|
| chr3_-_26060702 | 0.02 |
ENSDART00000144726
|
ypel3
|
yippee-like 3 |
| chr4_-_9779763 | 0.02 |
ENSDART00000134280
|
svopl
|
SVOP-like |
| chr9_-_30997261 | 0.02 |
ENSDART00000146300
|
klf5b
|
Kruppel-like factor 5b |
| chr13_-_22776819 | 0.01 |
ENSDART00000143097
|
rufy2
|
RUN and FYVE domain containing 2 |
| chr11_+_25063488 | 0.01 |
|
|
|
| chr13_-_37002066 | 0.01 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr1_+_58608185 | 0.01 |
ENSDART00000161872
ENSDART00000160658 |
parn
|
poly(A)-specific ribonuclease (deadenylation nuclease) |
| chr2_-_5811839 | 0.01 |
ENSDART00000100924
|
cwc25
|
CWC25 spliceosome-associated protein homolog (S. cerevisiae) |
| chr2_-_14758209 | 0.01 |
ENSDART00000162816
|
BX510915.1
|
ENSDARG00000099630 |
| chr5_-_34856057 | 0.01 |
ENSDART00000051312
|
ttc33
|
tetratricopeptide repeat domain 33 |
| chr7_+_39812522 | 0.01 |
ENSDART00000099046
|
ENSDARG00000036489
|
ENSDARG00000036489 |
| chr15_-_11798324 | 0.01 |
|
|
|
| chr13_+_33578737 | 0.01 |
ENSDART00000161465
|
CABZ01087953.1
|
ENSDARG00000104106 |
| chr18_+_9413588 | 0.01 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr15_+_5124897 | 0.01 |
ENSDART00000101937
|
pgm2l1
|
phosphoglucomutase 2-like 1 |
| chr21_+_15613081 | 0.01 |
ENSDART00000065772
|
ddt
|
D-dopachrome tautomerase |
| chr18_-_20605032 | 0.01 |
ENSDART00000134722
|
bcl2l13
|
BCL2-like 13 (apoptosis facilitator) |
| chr21_-_17001108 | 0.00 |
ENSDART00000101263
|
ube2g1b
|
ubiquitin-conjugating enzyme E2G 1b (UBC7 homolog, yeast) |
| chr7_+_48532749 | 0.00 |
ENSDART00000145375
|
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0050886 | sleep(GO:0030431) endocrine process(GO:0050886) |
| 0.1 | 0.3 | GO:0046166 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.6 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.3 | GO:1901492 | regulation of lymphangiogenesis(GO:1901490) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.1 | 0.3 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.2 | GO:0006531 | aspartate metabolic process(GO:0006531) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.4 | GO:0021767 | mammillary body development(GO:0021767) |
| 0.0 | 0.2 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.1 | GO:1903010 | regulation of bone development(GO:1903010) |
| 0.0 | 0.2 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) positive regulation of potassium ion transport(GO:0043268) positive regulation of potassium ion transmembrane transport(GO:1901381) positive regulation of sodium ion transmembrane transport(GO:1902307) regulation of potassium ion import(GO:1903286) positive regulation of potassium ion import(GO:1903288) positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0071236 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.3 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.1 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.2 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.4 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.3 | GO:0050773 | regulation of dendrite development(GO:0050773) |
| 0.0 | 0.1 | GO:0006798 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.0 | 0.2 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 0.2 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.4 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.3 | GO:0008929 | triose-phosphate isomerase activity(GO:0004807) methylglyoxal synthase activity(GO:0008929) |
| 0.1 | 0.6 | GO:0070095 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.2 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.2 | GO:0030151 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) molybdenum ion binding(GO:0030151) |
| 0.0 | 0.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.2 | GO:0017081 | chloride channel regulator activity(GO:0017081) chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.0 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.2 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.3 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0051430 | mu-type opioid receptor binding(GO:0031852) corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |