Project
DANIO-CODE
Navigation
Downloads
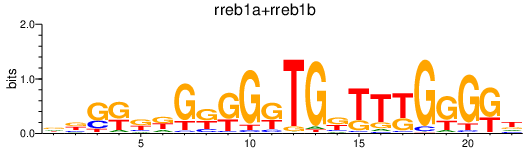
Results for rreb1a+rreb1b
Z-value: 1.03
Transcription factors associated with rreb1a+rreb1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rreb1b
|
ENSDARG00000042652 | ras responsive element binding protein 1b |
|
rreb1a
|
ENSDARG00000063701 | ras responsive element binding protein 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rreb1b | dr10_dc_chr2_+_21342233_21342398 | 0.74 | 1.1e-03 | Click! |
| rreb1a | dr10_dc_chr24_-_2453890_2453929 | -0.71 | 2.2e-03 | Click! |
Activity profile of rreb1a+rreb1b motif
Sorted Z-values of rreb1a+rreb1b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of rreb1a+rreb1b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_54171992 | 3.11 |
ENSDART00000122692
|
CABZ01112317.1
|
ENSDARG00000086057 |
| chr22_+_37944019 | 2.78 |
|
|
|
| chr13_-_508202 | 2.59 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr19_-_5441932 | 2.40 |
ENSDART00000105034
|
cyt1l
|
type I cytokeratin, enveloping layer, like |
| KN150170v1_+_10738 | 2.36 |
ENSDART00000159076
|
HUS1
|
HUS1 checkpoint clamp component |
| chr23_-_9833729 | 2.35 |
ENSDART00000045126
|
lama5
|
laminin, alpha 5 |
| chr1_-_4845236 | 2.26 |
ENSDART00000103755
ENSDART00000023692 |
fn1b
|
fibronectin 1b |
| chr13_-_508050 | 2.15 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr12_+_42281025 | 2.11 |
ENSDART00000167324
|
ebf3a
|
early B-cell factor 3a |
| chr12_+_2642541 | 2.08 |
ENSDART00000093113
|
antxr1c
|
anthrax toxin receptor 1c |
| chr5_-_71229872 | 1.98 |
|
|
|
| chr18_+_284399 | 1.89 |
ENSDART00000158040
|
larp6
|
La ribonucleoprotein domain family, member 6 |
| chr21_+_28408329 | 1.85 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr4_+_15870605 | 1.84 |
ENSDART00000178674
|
BX004864.1
|
ENSDARG00000107641 |
| chr13_-_11404389 | 1.77 |
ENSDART00000018155
|
adss
|
adenylosuccinate synthase |
| chr6_-_22504772 | 1.73 |
ENSDART00000170039
|
sept9b
|
septin 9b |
| KN149816v1_-_3337 | 1.68 |
|
|
|
| chr25_+_34270560 | 1.66 |
ENSDART00000146669
|
wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr11_+_44809527 | 1.65 |
ENSDART00000037819
ENSDART00000159886 |
tmc6b
|
transmembrane channel-like 6b |
| chr23_+_2101659 | 1.63 |
|
|
|
| chr21_-_44013995 | 1.61 |
ENSDART00000028960
|
ndufa2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 |
| chr21_-_43020159 | 1.51 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr14_-_9216303 | 1.50 |
ENSDART00000054689
|
atoh8
|
atonal bHLH transcription factor 8 |
| chr19_+_48499602 | 1.49 |
|
|
|
| chr8_-_1904720 | 1.47 |
ENSDART00000081563
|
si:dkey-178e17.3
|
si:dkey-178e17.3 |
| chr9_-_34146631 | 1.46 |
ENSDART00000177897
|
AL590134.2
|
ENSDARG00000107869 |
| chr9_+_30483319 | 1.45 |
ENSDART00000148148
|
srpx
|
sushi-repeat containing protein, X-linked |
| chr9_+_21911860 | 1.44 |
ENSDART00000102021
|
sox1a
|
SRY (sex determining region Y)-box 1a |
| chr9_+_37136519 | 1.43 |
ENSDART00000135281
ENSDART00000113562 |
si:dkey-3d4.3
|
si:dkey-3d4.3 |
| chr19_+_31046291 | 1.43 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr16_-_55320569 | 1.41 |
ENSDART00000156368
|
ENSDARG00000069583
|
ENSDARG00000069583 |
| chr2_-_44330405 | 1.40 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr17_+_8166167 | 1.40 |
ENSDART00000169900
|
cdc42bpaa
|
CDC42 binding protein kinase alpha (DMPK-like) a |
| chr25_+_11317025 | 1.38 |
|
|
|
| chr7_-_2230502 | 1.38 |
ENSDART00000173879
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr8_+_4311821 | 1.36 |
ENSDART00000015214
|
pptc7a
|
PTC7 protein phosphatase homolog a |
| chr19_-_5464238 | 1.36 |
ENSDART00000105806
|
cyt1
|
type I cytokeratin, enveloping layer |
| chr19_+_43414160 | 1.33 |
ENSDART00000033724
|
fabp3
|
fatty acid binding protein 3, muscle and heart |
| chr11_-_4216204 | 1.29 |
ENSDART00000121716
|
ENSDARG00000086300
|
ENSDARG00000086300 |
| chr16_-_39181608 | 1.28 |
ENSDART00000075517
|
gdf6a
|
growth differentiation factor 6a |
| chr5_-_71340996 | 1.27 |
ENSDART00000162526
|
smyd1a
|
SET and MYND domain containing 1a |
| chr6_+_59788008 | 1.27 |
|
|
|
| chr7_-_73530653 | 1.26 |
ENSDART00000166633
ENSDART00000009888 ENSDART00000171254 |
casq1b
|
calsequestrin 1b |
| KN150596v1_+_18342 | 1.21 |
ENSDART00000169155
|
ppic
|
peptidylprolyl isomerase C |
| chr10_+_22802607 | 1.20 |
ENSDART00000144845
|
tmem88a
|
transmembrane protein 88 a |
| chr11_-_25803101 | 1.18 |
ENSDART00000088888
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr17_+_1610578 | 1.18 |
ENSDART00000082101
|
ppp2r5ca
|
protein phosphatase 2, regulatory subunit B', gamma a |
| chr25_-_36886647 | 1.15 |
ENSDART00000160688
|
psma1
|
proteasome subunit alpha 1 |
| chr13_-_20387917 | 1.14 |
ENSDART00000167916
ENSDART00000165310 ENSDART00000168955 |
gfra1a
|
gdnf family receptor alpha 1a |
| chr7_-_12656353 | 1.13 |
ENSDART00000023301
ENSDART00000172901 ENSDART00000163045 ENSDART00000163461 |
sh3gl3a
|
SH3-domain GRB2-like 3a |
| chr4_-_16356071 | 1.12 |
ENSDART00000079521
|
kera
|
keratocan |
| chr25_+_5162045 | 1.12 |
ENSDART00000169540
|
ENSDARG00000101164
|
ENSDARG00000101164 |
| chr7_+_31567166 | 1.12 |
ENSDART00000099785
ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr3_+_16115708 | 1.10 |
ENSDART00000122519
|
st8sia6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
| chr13_-_25067585 | 1.10 |
ENSDART00000159585
|
adka
|
adenosine kinase a |
| chr3_-_38550961 | 1.09 |
ENSDART00000155042
|
mpp3a
|
membrane protein, palmitoylated 3a (MAGUK p55 subfamily member 3) |
| chr14_+_34174018 | 1.09 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr5_-_71643185 | 1.09 |
ENSDART00000029014
|
pax8
|
paired box 8 |
| chr16_+_5470544 | 1.08 |
|
|
|
| chr25_-_207828 | 1.08 |
ENSDART00000169152
|
srpk2
|
SRSF protein kinase 2 |
| chr15_+_37204488 | 1.07 |
ENSDART00000157762
|
aplp1
|
amyloid beta (A4) precursor-like protein 1 |
| chr13_-_31304614 | 1.07 |
|
|
|
| chr6_-_53426323 | 1.01 |
ENSDART00000162791
|
mst1
|
macrophage stimulating 1 |
| chr8_+_166461 | 1.00 |
|
|
|
| chr15_-_40328793 | 0.99 |
ENSDART00000155018
|
si:ch211-281l24.3
|
si:ch211-281l24.3 |
| chr23_+_45020761 | 0.99 |
ENSDART00000159104
|
atp1b2a
|
ATPase, Na+/K+ transporting, beta 2a polypeptide |
| chr11_+_23522743 | 0.98 |
ENSDART00000121874
|
nfasca
|
neurofascin homolog (chicken) a |
| chr25_+_57432 | 0.95 |
ENSDART00000128281
|
cntn1a
|
contactin 1a |
| chr3_+_59716594 | 0.95 |
ENSDART00000157351
ENSDART00000153928 |
si:ch211-110e21.3
|
si:ch211-110e21.3 |
| chr16_+_22672315 | 0.94 |
ENSDART00000142241
|
she
|
Src homology 2 domain containing E |
| chr25_+_17366098 | 0.94 |
|
|
|
| chr23_+_22350713 | 0.94 |
ENSDART00000130375
|
rap1gap
|
RAP1 GTPase activating protein |
| chr19_-_6856033 | 0.94 |
ENSDART00000170952
|
pvrl2l
|
poliovirus receptor-related 2 like |
| chr12_+_41016238 | 0.93 |
ENSDART00000170976
|
kif5bb
|
kinesin family member 5B, b |
| chr12_+_1370058 | 0.93 |
ENSDART00000026303
|
rasd1
|
RAS, dexamethasone-induced 1 |
| chr6_+_59762717 | 0.93 |
ENSDART00000050457
|
zgc:65895
|
zgc:65895 |
| chr14_-_46646747 | 0.92 |
|
|
|
| chr17_-_20830398 | 0.92 |
ENSDART00000149630
|
ank3a
|
ankyrin 3a |
| chr21_-_45573799 | 0.90 |
ENSDART00000158489
|
zgc:77058
|
zgc:77058 |
| chr25_+_5232865 | 0.90 |
|
|
|
| chr18_+_284567 | 0.90 |
ENSDART00000158040
|
larp6
|
La ribonucleoprotein domain family, member 6 |
| KN150039v1_-_936 | 0.89 |
ENSDART00000168136
|
HDGFL3
|
HDGF like 3 |
| chr19_-_9192541 | 0.88 |
|
|
|
| KN149973v1_+_28300 | 0.88 |
|
|
|
| chr15_-_16271208 | 0.88 |
|
|
|
| chr18_+_30391910 | 0.88 |
ENSDART00000158871
|
gse1
|
Gse1 coiled-coil protein |
| chr25_+_36878565 | 0.87 |
ENSDART00000157596
|
ric3b
|
RIC3 acetylcholine receptor chaperone b |
| chr5_-_1498277 | 0.87 |
|
|
|
| chr17_+_53224434 | 0.87 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr5_-_70915664 | 0.86 |
|
|
|
| chr5_+_474163 | 0.86 |
ENSDART00000055681
|
tek
|
TEK tyrosine kinase, endothelial |
| chr10_+_24476213 | 0.85 |
ENSDART00000146370
|
slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr21_-_41445163 | 0.85 |
ENSDART00000170457
|
hmp19
|
HMP19 protein |
| chr12_+_13206107 | 0.85 |
ENSDART00000105896
|
atp2a1l
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 like |
| chr9_-_216905 | 0.84 |
ENSDART00000163068
|
aaas
|
achalasia, adrenocortical insufficiency, alacrimia |
| chr10_-_43466392 | 0.84 |
ENSDART00000062631
|
hapln1b
|
hyaluronan and proteoglycan link protein 1b |
| chr22_-_29577 | 0.82 |
ENSDART00000158058
|
NBL1
|
MINOS1-NBL1 readthrough |
| chr10_-_43466478 | 0.81 |
ENSDART00000062631
|
hapln1b
|
hyaluronan and proteoglycan link protein 1b |
| chr17_+_52736192 | 0.81 |
ENSDART00000158273
ENSDART00000161414 |
meis2a
|
Meis homeobox 2a |
| chr13_+_22545318 | 0.81 |
ENSDART00000143312
|
zgc:193505
|
zgc:193505 |
| KN149968v1_+_15749 | 0.80 |
ENSDART00000168977
|
ENSDARG00000102503
|
ENSDARG00000102503 |
| chr20_-_35343242 | 0.80 |
ENSDART00000113294
|
fzd3a
|
frizzled class receptor 3a |
| chr21_+_25729090 | 0.80 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr9_-_46272322 | 0.79 |
ENSDART00000169682
|
hdac4
|
histone deacetylase 4 |
| chr2_+_23038657 | 0.78 |
ENSDART00000089012
|
kif1ab
|
kinesin family member 1Ab |
| chr24_+_26944182 | 0.77 |
ENSDART00000089541
|
dip2ca
|
disco-interacting protein 2 homolog Ca |
| KN149987v1_-_29629 | 0.77 |
ENSDART00000159555
ENSDART00000168161 |
ENSDARG00000103311
|
ENSDARG00000103311 |
| chr9_+_22953351 | 0.76 |
ENSDART00000090875
|
tnfaip6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr14_-_46646451 | 0.76 |
|
|
|
| chr20_-_10133201 | 0.75 |
ENSDART00000144970
|
meis2b
|
Meis homeobox 2b |
| chr7_+_16677929 | 0.73 |
|
|
|
| chr25_+_33364224 | 0.73 |
ENSDART00000121449
|
roraa
|
RAR-related orphan receptor A, paralog a |
| chr8_+_388003 | 0.72 |
ENSDART00000067668
|
pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr25_+_7660590 | 0.72 |
ENSDART00000155016
|
dgkzb
|
diacylglycerol kinase, zeta b |
| chr19_-_47897712 | 0.72 |
|
|
|
| chr22_+_1169970 | 0.71 |
ENSDART00000171060
ENSDART00000167710 |
eps8l3a
|
EPS8-like 3a |
| chr22_-_95158 | 0.71 |
|
|
|
| chr17_-_20830287 | 0.71 |
ENSDART00000149630
|
ank3a
|
ankyrin 3a |
| chr24_-_2373152 | 0.70 |
|
|
|
| chr14_+_503643 | 0.70 |
|
|
|
| chr10_-_38372282 | 0.68 |
ENSDART00000149580
|
nrip1b
|
nuclear receptor interacting protein 1b |
| chr10_-_45535333 | 0.68 |
|
|
|
| chr17_-_5703094 | 0.66 |
ENSDART00000058894
|
si:ch73-340m8.2
|
si:ch73-340m8.2 |
| chr9_+_1225808 | 0.66 |
ENSDART00000177730
|
CABZ01114359.1
|
ENSDARG00000107255 |
| chr21_+_45693978 | 0.66 |
ENSDART00000160324
|
CABZ01102047.1
|
ENSDARG00000099648 |
| KN150487v1_+_17905 | 0.65 |
|
|
|
| chr23_-_45250886 | 0.65 |
ENSDART00000163367
|
ENSDARG00000103727
|
ENSDARG00000103727 |
| chr6_+_54231519 | 0.64 |
ENSDART00000149542
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr5_-_337100 | 0.64 |
|
|
|
| chr2_+_53253623 | 0.63 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr23_-_1124499 | 0.63 |
|
|
|
| chr13_-_45386047 | 0.62 |
ENSDART00000137566
|
rhd
|
Rh blood group, D antigen |
| KN150233v1_-_1233 | 0.62 |
|
|
|
| chr14_+_51848088 | 0.61 |
ENSDART00000168874
|
rpl26
|
ribosomal protein L26 |
| chr12_+_10015987 | 0.61 |
ENSDART00000078497
|
slc4a1b
|
solute carrier family 4 (anion exchanger), member 1b (Diego blood group) |
| chr12_+_7421078 | 0.60 |
ENSDART00000163114
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr16_+_10974655 | 0.60 |
|
|
|
| chr7_+_34394526 | 0.59 |
|
|
|
| chr11_+_18817797 | 0.58 |
|
|
|
| chr22_+_980836 | 0.57 |
ENSDART00000149743
|
pparda
|
peroxisome proliferator-activated receptor delta a |
| chr11_+_44035254 | 0.56 |
|
|
|
| chr6_+_55091896 | 0.56 |
|
|
|
| KN150503v1_-_598 | 0.56 |
ENSDART00000174608
|
CABZ01110189.1
|
ENSDARG00000108333 |
| chr21_-_36495488 | 0.55 |
|
|
|
| chr2_-_44893769 | 0.54 |
ENSDART00000041806
|
acsm3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr3_+_1455242 | 0.54 |
ENSDART00000065922
|
wbp2nl
|
WBP2 N-terminal like |
| chr21_+_5293086 | 0.54 |
|
|
|
| chr14_+_151136 | 0.54 |
ENSDART00000165766
|
mcm7
|
minichromosome maintenance complex component 7 |
| chr16_-_12579115 | 0.54 |
ENSDART00000147483
|
ephb6
|
eph receptor B6 |
| chr12_+_49091872 | 0.53 |
ENSDART00000178771
|
CABZ01075125.4
|
ENSDARG00000107503 |
| chr19_-_5448129 | 0.53 |
ENSDART00000004812
|
cyt1
|
type I cytokeratin, enveloping layer |
| KN150411v1_-_12184 | 0.52 |
|
|
|
| chr8_+_54088321 | 0.52 |
|
|
|
| chr12_+_42281154 | 0.52 |
ENSDART00000167324
|
ebf3a
|
early B-cell factor 3a |
| chr23_+_4440723 | 0.51 |
|
|
|
| chr21_+_44586326 | 0.51 |
|
|
|
| chr15_+_18710214 | 0.50 |
ENSDART00000166555
|
cadm1b
|
cell adhesion molecule 1b |
| chr6_-_43679553 | 0.50 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr2_+_2259793 | 0.49 |
ENSDART00000109073
|
dsg2l
|
desmoglein 2 like |
| chr16_+_6693574 | 0.49 |
|
|
|
| chr14_+_23296676 | 0.47 |
ENSDART00000159573
|
med12
|
mediator complex subunit 12 |
| chr25_-_4296732 | 0.47 |
|
|
|
| chr23_-_339083 | 0.47 |
ENSDART00000140986
|
uhrf1bp1
|
UHRF1 binding protein 1 |
| KN150423v1_-_9765 | 0.46 |
|
|
|
| chr10_-_36882126 | 0.46 |
ENSDART00000111104
|
phf12a
|
PHD finger protein 12a |
| chr9_+_19584625 | 0.45 |
ENSDART00000168357
|
si:dkey-281i8.1
|
si:dkey-281i8.1 |
| chr18_+_30392138 | 0.45 |
ENSDART00000165450
|
gse1
|
Gse1 coiled-coil protein |
| chr18_+_21133694 | 0.45 |
ENSDART00000060015
|
chka
|
choline kinase alpha |
| chr14_-_46646556 | 0.44 |
|
|
|
| chr2_+_28906539 | 0.44 |
ENSDART00000078232
|
cdh10a
|
cadherin 10, type 2a (T2-cadherin) |
| chr8_-_387982 | 0.43 |
|
|
|
| chr15_-_3120500 | 0.43 |
|
|
|
| chr12_-_1931281 | 0.43 |
ENSDART00000005676
ENSDART00000127937 |
sox9a
|
SRY (sex determining region Y)-box 9a |
| chr16_+_11027394 | 0.43 |
ENSDART00000172949
|
pou2f2a
|
POU class 2 homeobox 2a |
| chr12_-_28248133 | 0.43 |
ENSDART00000016283
|
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr23_-_7048903 | 0.43 |
ENSDART00000149656
|
edem2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr12_-_41128584 | 0.42 |
|
|
|
| chr20_-_7141247 | 0.42 |
ENSDART00000160910
|
si:ch211-121a2.4
|
si:ch211-121a2.4 |
| chr16_+_5470419 | 0.41 |
|
|
|
| chr23_-_4919504 | 0.41 |
ENSDART00000133701
|
slc6a1a
|
solute carrier family 6 (neurotransmitter transporter), member 1a |
| chr10_+_33956052 | 0.40 |
|
|
|
| chr3_+_23573114 | 0.40 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr5_+_30880165 | 0.40 |
ENSDART00000098197
|
ENSDARG00000035471
|
ENSDARG00000035471 |
| chr5_+_1164747 | 0.40 |
ENSDART00000129490
|
gb:bc139872
|
expressed sequence BC139872 |
| chr1_+_51712192 | 0.40 |
|
|
|
| chr10_+_213828 | 0.38 |
ENSDART00000138812
|
mpzl1l
|
myelin protein zero-like 1 like |
| chr17_+_23102259 | 0.38 |
ENSDART00000178403
|
rasgrp3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr21_+_19797559 | 0.37 |
ENSDART00000147555
|
ccdc80l2
|
coiled-coil domain containing 80 like 2 |
| chr21_+_25588388 | 0.37 |
ENSDART00000134678
|
ENSDARG00000078256
|
ENSDARG00000078256 |
| chr21_+_8334453 | 0.37 |
ENSDART00000055336
|
dennd1a
|
DENN/MADD domain containing 1A |
| chr16_+_54729975 | 0.37 |
|
|
|
| chr21_+_19797435 | 0.36 |
ENSDART00000147555
|
ccdc80l2
|
coiled-coil domain containing 80 like 2 |
| chr16_+_25845432 | 0.36 |
|
|
|
| chr11_-_3888456 | 0.35 |
ENSDART00000175310
ENSDART00000082404 |
rpn1
|
ribophorin I |
| chr10_-_45212226 | 0.35 |
ENSDART00000170413
|
mrps24
|
mitochondrial ribosomal protein S24 |
| chr1_-_51862897 | 0.34 |
ENSDART00000136469
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr16_+_68014 | 0.33 |
ENSDART00000159652
|
sox4b
|
SRY (sex determining region Y)-box 4b |
| KN149797v1_+_49 | 0.33 |
ENSDART00000176987
|
CABZ01113190.1
|
ENSDARG00000107708 |
| chr23_+_11734349 | 0.33 |
ENSDART00000091416
|
cntn3a.1
|
contactin 3a, tandem duplicate 1 |
| chr6_-_43679476 | 0.33 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.5 | 2.3 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.4 | 1.3 | GO:0043703 | blood vessel maturation(GO:0001955) retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.4 | 1.3 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.4 | 1.2 | GO:2000725 | cardiac cell fate commitment(GO:0060911) regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.4 | 1.5 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.4 | 1.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.3 | 5.1 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.3 | 1.6 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.3 | 1.5 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.3 | 1.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.3 | 0.8 | GO:0036073 | perichondral ossification(GO:0036073) |
| 0.3 | 1.0 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.2 | 0.7 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.2 | 0.6 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 2.3 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.2 | 1.1 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.2 | 1.9 | GO:0009251 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.1 | 0.9 | GO:0010595 | positive regulation of endothelial cell migration(GO:0010595) |
| 0.1 | 1.7 | GO:0060021 | palate development(GO:0060021) |
| 0.1 | 0.1 | GO:0010885 | cholesterol storage(GO:0010878) regulation of cholesterol storage(GO:0010885) |
| 0.1 | 1.1 | GO:0072098 | anterior/posterior pattern specification involved in pronephros development(GO:0034672) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.1 | 0.2 | GO:0003211 | cardiac ventricle formation(GO:0003211) |
| 0.1 | 0.5 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.1 | 1.2 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.1 | 1.3 | GO:0030241 | skeletal muscle thin filament assembly(GO:0030240) skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 1.0 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.1 | 0.6 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.6 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 1.0 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.1 | 0.7 | GO:0003209 | cardiac atrium morphogenesis(GO:0003209) |
| 0.1 | 0.5 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 0.5 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.7 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 0.4 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.1 | 0.9 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.1 | 0.3 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.1 | 1.0 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 1.2 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 1.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.5 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.9 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.3 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 1.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.5 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.0 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 0.8 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 0.4 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 1.0 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.9 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 0.3 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 1.7 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.1 | GO:0046636 | negative regulation of alpha-beta T cell activation(GO:0046636) negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.3 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.2 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 1.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.7 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.0 | 1.1 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.3 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.6 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.3 | GO:0070654 | sensory epithelium regeneration(GO:0070654) epithelium regeneration(GO:1990399) |
| 0.0 | 0.2 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.3 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 1.5 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 1.3 | GO:0001947 | heart looping(GO:0001947) |
| 0.0 | 1.7 | GO:0030833 | regulation of actin filament polymerization(GO:0030833) |
| 0.0 | 0.8 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.3 | GO:0032543 | mitochondrial translation(GO:0032543) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.2 | 2.3 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 1.0 | GO:1904949 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.1 | 1.6 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 1.3 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 1.7 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 2.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.7 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 0.3 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 4.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.4 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 1.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.5 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.2 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 1.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 1.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.5 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 1.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 1.8 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 7.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.8 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.4 | 1.7 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.4 | 1.9 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.4 | 1.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.3 | 2.3 | GO:0001948 | glycoprotein binding(GO:0001948) proteoglycan binding(GO:0043394) |
| 0.2 | 1.5 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 2.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.5 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.1 | 1.0 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.9 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 1.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 0.7 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 1.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.1 | 1.0 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 1.2 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 2.3 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.6 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 0.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.9 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.1 | 1.2 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 0.8 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.1 | 1.2 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 1.0 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.8 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.6 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.8 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.4 | GO:0015924 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.6 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.1 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.5 | GO:0015645 | fatty acid ligase activity(GO:0015645) |
| 0.0 | 0.7 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.3 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.2 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.5 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 1.3 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 0.8 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.7 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 5.9 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.5 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.1 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.0 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.7 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 0.8 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.1 | 1.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.9 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 2.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.9 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 2.9 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 0.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.1 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.8 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.1 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 1.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.4 | REACTOME DOWNSTREAM SIGNALING EVENTS OF B CELL RECEPTOR BCR | Genes involved in Downstream Signaling Events Of B Cell Receptor (BCR) |