Project
DANIO-CODE
Navigation
Downloads
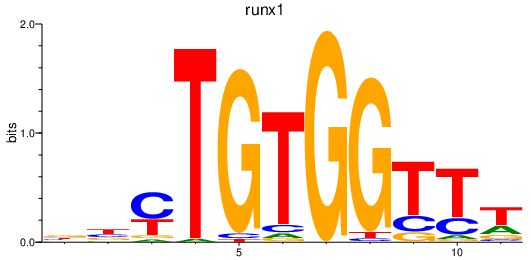
Results for runx1
Z-value: 1.10
Transcription factors associated with runx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
runx1
|
ENSDARG00000087646 | RUNX family transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| runx1 | dr10_dc_chr1_-_1254253_1254255 | 0.27 | 3.1e-01 | Click! |
Activity profile of runx1 motif
Sorted Z-values of runx1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of runx1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr25_+_222244 | 1.20 |
ENSDART00000155344
|
ENSDARG00000073905
|
ENSDARG00000073905 |
| chr2_-_16896965 | 1.12 |
ENSDART00000022549
|
atp1b3a
|
ATPase, Na+/K+ transporting, beta 3a polypeptide |
| chr8_-_38168395 | 1.02 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr23_-_3814291 | 0.92 |
|
|
|
| chr21_-_11539798 | 0.92 |
ENSDART00000144770
|
cast
|
calpastatin |
| chr8_-_37217197 | 0.91 |
ENSDART00000178556
|
rbm39b
|
RNA binding motif protein 39b |
| chr4_+_50923792 | 0.86 |
ENSDART00000161370
|
BX640547.3
|
ENSDARG00000100076 |
| chr2_+_36916494 | 0.85 |
ENSDART00000084859
|
rabgap1l2
|
RAB GTPase activating protein 1-like 2 |
| chr9_-_23307419 | 0.83 |
ENSDART00000020884
|
lypd6
|
LY6/PLAUR domain containing 6 |
| chr16_-_9978112 | 0.81 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr11_-_11591394 | 0.73 |
ENSDART00000142208
|
zgc:110712
|
zgc:110712 |
| chr11_+_5467173 | 0.71 |
|
|
|
| chr11_+_37639045 | 0.71 |
ENSDART00000111157
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr8_+_20125687 | 0.70 |
ENSDART00000124809
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr25_+_5232865 | 0.70 |
|
|
|
| chr16_-_45943282 | 0.69 |
ENSDART00000133213
|
afp4
|
antifreeze protein type IV |
| chr15_-_26703761 | 0.69 |
ENSDART00000087632
|
ENSDARG00000079340
|
ENSDARG00000079340 |
| chr23_+_44839349 | 0.69 |
ENSDART00000143399
|
camta2
|
calmodulin binding transcription activator 2 |
| chr21_-_19280266 | 0.69 |
ENSDART00000141596
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr16_-_17148579 | 0.67 |
ENSDART00000138715
|
plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr5_-_17542077 | 0.67 |
ENSDART00000144898
|
rnf215
|
ring finger protein 215 |
| chr1_-_50066633 | 0.67 |
|
|
|
| chr7_-_24093289 | 0.66 |
ENSDART00000064789
|
txn
|
thioredoxin |
| chr16_-_51390400 | 0.66 |
ENSDART00000148894
|
serpinb14
|
serpin peptidase inhibitor, clade B (ovalbumin), member 14 |
| chr18_+_22120282 | 0.65 |
ENSDART00000147230
|
zgc:158868
|
zgc:158868 |
| chr12_+_38382027 | 0.64 |
ENSDART00000021069
|
rpl38
|
ribosomal protein L38 |
| chr10_-_43924675 | 0.64 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr9_-_14533551 | 0.63 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr11_-_22450582 | 0.63 |
|
|
|
| chr1_-_11439671 | 0.63 |
ENSDART00000164817
|
mtp
|
microsomal triglyceride transfer protein |
| chr3_+_54506912 | 0.62 |
ENSDART00000135913
|
si:ch211-74m13.1
|
si:ch211-74m13.1 |
| chr13_-_42986821 | 0.60 |
ENSDART00000164439
|
si:ch211-106f21.1
|
si:ch211-106f21.1 |
| chr14_-_3264798 | 0.60 |
ENSDART00000171601
|
im:7150988
|
im:7150988 |
| chr25_+_5233045 | 0.59 |
|
|
|
| chr13_-_39033893 | 0.58 |
ENSDART00000045434
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr8_-_50992034 | 0.58 |
ENSDART00000004065
|
zgc:91909
|
zgc:91909 |
| chr2_-_6139895 | 0.58 |
ENSDART00000147509
|
si:ch211-284b7.3
|
si:ch211-284b7.3 |
| chr7_+_25758469 | 0.58 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr22_+_2921305 | 0.57 |
ENSDART00000143258
|
cep19
|
centrosomal protein 19 |
| chr7_-_48392300 | 0.57 |
|
|
|
| chr8_+_21321765 | 0.56 |
ENSDART00000131691
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr25_-_34645938 | 0.55 |
ENSDART00000108895
|
ENSDARG00000077587
|
ENSDARG00000077587 |
| chr6_-_7281412 | 0.55 |
ENSDART00000053776
|
fkbp11
|
FK506 binding protein 11 |
| chr21_+_34895120 | 0.55 |
ENSDART00000157904
ENSDART00000135806 |
si:dkey-71d15.2
|
si:dkey-71d15.2 |
| chr2_-_58822653 | 0.54 |
ENSDART00000114286
ENSDART00000114181 |
sugp1
|
SURP and G patch domain containing 1 |
| chr20_+_54289892 | 0.54 |
ENSDART00000060444
|
rps29
|
ribosomal protein S29 |
| chr3_+_3955069 | 0.54 |
ENSDART00000092393
|
plbd1
|
phospholipase B domain containing 1 |
| chr2_-_44429891 | 0.54 |
ENSDART00000163040
ENSDART00000056372 ENSDART00000109251 ENSDART00000166923 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr4_-_76620869 | 0.53 |
|
|
|
| chr13_-_36996246 | 0.53 |
ENSDART00000133242
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr5_+_60790602 | 0.52 |
ENSDART00000127521
|
lrwd1
|
leucine-rich repeats and WD repeat domain containing 1 |
| chr24_+_19374200 | 0.52 |
ENSDART00000056081
ENSDART00000027022 |
sulf1
|
sulfatase 1 |
| chr6_+_7092350 | 0.52 |
ENSDART00000049695
ENSDART00000136088 ENSDART00000083424 ENSDART00000125912 |
dzip1
|
DAZ interacting zinc finger protein 1 |
| chr3_+_37645350 | 0.51 |
ENSDART00000134524
|
rps11
|
ribosomal protein S11 |
| chr12_+_6007990 | 0.51 |
ENSDART00000091868
|
g6pca.2
|
glucose-6-phosphatase a, catalytic subunit, tandem duplicate 2 |
| chr15_+_34131126 | 0.51 |
ENSDART00000169487
|
vwde
|
von Willebrand factor D and EGF domains |
| chr14_-_2008649 | 0.50 |
ENSDART00000161817
|
pcdh2g16
|
protocadherin 2 gamma 16 |
| chr7_+_5372830 | 0.50 |
ENSDART00000135271
|
MYADM
|
myeloid associated differentiation marker |
| chr7_+_65532390 | 0.50 |
ENSDART00000156683
|
CT573494.2
|
ENSDARG00000097673 |
| chr2_-_3202918 | 0.50 |
|
|
|
| chr8_+_7301525 | 0.50 |
ENSDART00000092347
|
ssuh2rs1
|
ssu-2 homolog, related sequence 1 |
| chr4_-_72146177 | 0.50 |
ENSDART00000150546
|
si:dkey-262g12.3
|
si:dkey-262g12.3 |
| chr1_-_54331157 | 0.49 |
ENSDART00000083572
|
zgc:136864
|
zgc:136864 |
| chr6_+_30681170 | 0.49 |
ENSDART00000112294
|
ttc22
|
tetratricopeptide repeat domain 22 |
| chr1_-_11163480 | 0.49 |
ENSDART00000033361
|
ttyh3b
|
tweety family member 3b |
| chr14_+_38445969 | 0.49 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr7_-_6296880 | 0.49 |
ENSDART00000172954
|
si:ch1073-153i20.4
|
si:ch1073-153i20.4 |
| chr12_+_16403743 | 0.48 |
ENSDART00000058665
|
kif20bb
|
kinesin family member 20Bb |
| chr17_+_24613874 | 0.48 |
ENSDART00000093004
|
map4k3b
|
mitogen-activated protein kinase kinase kinase kinase 3b |
| chr12_+_31558667 | 0.47 |
ENSDART00000152971
|
dnmbp
|
dynamin binding protein |
| chr2_+_1767497 | 0.47 |
ENSDART00000029371
|
dspa
|
desmoplakin a |
| chr14_+_47229400 | 0.47 |
ENSDART00000136045
|
mgst2
|
microsomal glutathione S-transferase 2 |
| chr8_+_32604349 | 0.47 |
ENSDART00000126833
|
hmcn2
|
hemicentin 2 |
| chr11_-_471166 | 0.46 |
ENSDART00000154888
|
cnbpb
|
CCHC-type zinc finger, nucleic acid binding protein b |
| chr6_+_56163589 | 0.46 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr11_+_7570027 | 0.45 |
ENSDART00000091550
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr13_-_5953330 | 0.45 |
ENSDART00000099224
|
dld
|
deltaD |
| chr18_+_40364675 | 0.44 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr12_-_26760324 | 0.44 |
ENSDART00000047724
|
zeb1b
|
zinc finger E-box binding homeobox 1b |
| chr20_+_52653111 | 0.44 |
ENSDART00000057980
|
tsta3
|
tissue specific transplantation antigen P35B |
| chr16_-_51390484 | 0.43 |
ENSDART00000148894
|
serpinb14
|
serpin peptidase inhibitor, clade B (ovalbumin), member 14 |
| chr2_-_9950224 | 0.43 |
ENSDART00000146407
|
si:ch1073-170o4.1
|
si:ch1073-170o4.1 |
| chr16_-_29593569 | 0.43 |
ENSDART00000150028
|
onecutl
|
one cut domain, family member, like |
| chr17_-_42293039 | 0.42 |
ENSDART00000014296
|
foxa2
|
forkhead box A2 |
| chr22_-_613948 | 0.42 |
ENSDART00000148692
|
apobec2a
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2a |
| chr11_-_40415291 | 0.42 |
|
|
|
| chr10_+_36254196 | 0.42 |
ENSDART00000114102
|
COA4
|
cytochrome c oxidase assembly factor 4 homolog |
| chr6_+_45917081 | 0.41 |
ENSDART00000149450
ENSDART00000149642 |
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr16_+_4288700 | 0.41 |
ENSDART00000159694
|
inpp5b
|
inositol polyphosphate-5-phosphatase B |
| chr11_+_7570097 | 0.41 |
ENSDART00000091550
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr4_-_12796590 | 0.41 |
ENSDART00000075127
|
b2m
|
beta-2-microglobulin |
| chr17_+_15425559 | 0.41 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr1_-_24486146 | 0.41 |
ENSDART00000144711
|
tmem154
|
transmembrane protein 154 |
| chr11_-_42096921 | 0.40 |
|
|
|
| chr23_+_42396834 | 0.40 |
ENSDART00000164249
|
cyp2aa4
|
cytochrome P450, family 2, subfamily AA, polypeptide 4 |
| chr15_-_1061845 | 0.40 |
ENSDART00000156003
|
znf1010
|
zinc finger protein 1010 |
| chr12_-_5383331 | 0.40 |
ENSDART00000152622
ENSDART00000028043 |
noc3l
|
NOC3-like DNA replication regulator |
| chr19_+_1356824 | 0.40 |
ENSDART00000081779
|
zdhhc3b
|
zinc finger, DHHC-type containing 3b |
| chr10_+_33227967 | 0.39 |
ENSDART00000159666
|
myl10
|
myosin, light chain 10, regulatory |
| chr7_-_26153788 | 0.39 |
|
|
|
| chr10_-_8094671 | 0.39 |
ENSDART00000099033
|
zgc:158494
|
zgc:158494 |
| chr20_-_14158114 | 0.39 |
ENSDART00000009549
|
rhag
|
Rh-associated glycoprotein |
| chr9_-_216905 | 0.39 |
ENSDART00000163068
|
aaas
|
achalasia, adrenocortical insufficiency, alacrimia |
| chr20_+_32503118 | 0.39 |
ENSDART00000018640
|
snx3
|
sorting nexin 3 |
| chr9_-_56292487 | 0.38 |
ENSDART00000151720
|
si:ch211-39i22.1
|
si:ch211-39i22.1 |
| chr4_+_29817277 | 0.38 |
ENSDART00000167186
|
BX323023.2
|
ENSDARG00000104750 |
| chr21_-_2143830 | 0.38 |
|
|
|
| KN149817v1_+_2207 | 0.38 |
|
|
|
| chr4_-_2595775 | 0.38 |
ENSDART00000132581
ENSDART00000019508 |
csrp2
|
cysteine and glycine-rich protein 2 |
| chr6_-_43273456 | 0.38 |
|
|
|
| chr8_-_16662185 | 0.38 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr22_-_3264630 | 0.38 |
ENSDART00000170992
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr8_+_10267246 | 0.37 |
ENSDART00000159312
ENSDART00000160766 |
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr18_+_27753985 | 0.37 |
ENSDART00000141835
|
tspan18b
|
tetraspanin 18b |
| chr5_+_35839198 | 0.37 |
ENSDART00000102973
ENSDART00000103020 |
eda
|
ectodysplasin A |
| chr24_+_19374099 | 0.37 |
ENSDART00000056081
ENSDART00000027022 |
sulf1
|
sulfatase 1 |
| chr11_-_11347349 | 0.37 |
ENSDART00000173019
|
mrpl4
|
mitochondrial ribosomal protein L4 |
| chr20_-_47123598 | 0.36 |
ENSDART00000100320
|
dnmt3aa
|
DNA (cytosine-5-)-methyltransferase 3 alpha a |
| chr20_+_34640226 | 0.36 |
ENSDART00000076946
|
ENSDARG00000054723
|
ENSDARG00000054723 |
| chr23_+_8671759 | 0.36 |
ENSDART00000129997
|
rgs19
|
regulator of G protein signaling 19 |
| chr24_-_37800277 | 0.36 |
ENSDART00000141414
|
zgc:112185
|
zgc:112185 |
| chr7_-_6295302 | 0.36 |
ENSDART00000173199
|
HIST2H2AB (1 of many)
|
si:ch1073-153i20.5 |
| chr9_-_11589126 | 0.36 |
ENSDART00000146832
|
cryba2b
|
crystallin, beta A2b |
| chr7_-_6273677 | 0.36 |
ENSDART00000173419
|
si:ch73-368j24.1
|
si:ch73-368j24.1 |
| chr22_-_2921219 | 0.36 |
ENSDART00000092991
|
pigx
|
phosphatidylinositol glycan anchor biosynthesis, class X |
| chr12_+_3043444 | 0.36 |
ENSDART00000149427
|
sgca
|
sarcoglycan, alpha |
| chr3_-_19218660 | 0.35 |
ENSDART00000132987
|
s1pr5a
|
sphingosine-1-phosphate receptor 5a |
| chr15_-_15532980 | 0.35 |
ENSDART00000004220
ENSDART00000131259 |
rab34a
|
RAB34, member RAS oncogene family a |
| chr5_+_34022151 | 0.35 |
ENSDART00000141338
|
enc1
|
ectodermal-neural cortex 1 |
| chr14_-_31184501 | 0.35 |
|
|
|
| chr14_+_24548729 | 0.35 |
|
|
|
| chr23_+_36208816 | 0.35 |
ENSDART00000127174
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr3_+_27582726 | 0.35 |
ENSDART00000019004
|
arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma |
| chr4_+_72347854 | 0.35 |
ENSDART00000174148
|
CABZ01069015.2
|
ENSDARG00000105790 |
| chr19_+_7576862 | 0.34 |
ENSDART00000010862
|
mrpl24
|
mitochondrial ribosomal protein L24 |
| chr15_-_35394535 | 0.34 |
ENSDART00000144153
|
mff
|
mitochondrial fission factor |
| chr7_+_5790032 | 0.34 |
ENSDART00000173025
|
si:dkey-23a13.9
|
si:dkey-23a13.9 |
| chr5_+_23092679 | 0.34 |
|
|
|
| chr8_-_41266057 | 0.34 |
ENSDART00000108518
|
rnf10
|
ring finger protein 10 |
| chr22_-_583831 | 0.34 |
ENSDART00000106636
|
srsf3b
|
serine/arginine-rich splicing factor 3b |
| chr23_+_27052594 | 0.34 |
ENSDART00000109712
ENSDART00000018654 |
rnd1b
|
Rho family GTPase 1b |
| chr15_-_20254259 | 0.34 |
ENSDART00000157149
|
exoc3l2b
|
exocyst complex component 3-like 2b |
| chr5_+_23093316 | 0.34 |
ENSDART00000006983
|
kat5a
|
K(lysine) acetyltransferase 5a |
| chr20_+_4512872 | 0.34 |
|
|
|
| chr7_-_11812634 | 0.34 |
ENSDART00000101537
|
mex3b
|
mex-3 RNA binding family member B |
| chr5_-_22090338 | 0.33 |
|
|
|
| chr5_+_3965645 | 0.33 |
ENSDART00000100061
|
prdx4
|
peroxiredoxin 4 |
| chr14_-_26138828 | 0.33 |
ENSDART00000140173
|
si:dkeyp-110e4.6
|
si:dkeyp-110e4.6 |
| chr5_+_69369417 | 0.33 |
|
|
|
| chr21_-_14729288 | 0.33 |
ENSDART00000067001
|
noc4l
|
nucleolar complex associated 4 homolog |
| chr21_+_22808694 | 0.33 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr23_+_23730717 | 0.32 |
|
|
|
| chr7_+_20215723 | 0.32 |
ENSDART00000173724
ENSDART00000173773 |
si:dkey-33c9.8
|
si:dkey-33c9.8 |
| chr19_-_9192541 | 0.32 |
|
|
|
| chr23_-_29138952 | 0.32 |
ENSDART00000002812
|
casz1
|
castor zinc finger 1 |
| chr6_+_59788008 | 0.32 |
|
|
|
| chr14_+_37206949 | 0.32 |
|
|
|
| chr13_+_24132617 | 0.32 |
ENSDART00000135992
|
abcb10
|
ATP-binding cassette, sub-family B (MDR/TAP), member 10 |
| chr3_+_22962233 | 0.32 |
ENSDART00000142884
|
gngt2a
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2a |
| chr13_-_39821399 | 0.32 |
ENSDART00000056996
|
sfrp5
|
secreted frizzled-related protein 5 |
| chr23_-_4990023 | 0.32 |
ENSDART00000142699
|
taz
|
tafazzin |
| chr2_+_48052054 | 0.31 |
ENSDART00000159701
|
ftr23
|
finTRIM family, member 23 |
| chr6_-_34955205 | 0.31 |
ENSDART00000131610
|
serbp1a
|
SERPINE1 mRNA binding protein 1a |
| chr5_-_20375703 | 0.31 |
ENSDART00000134697
|
pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr1_+_32485317 | 0.31 |
ENSDART00000136761
|
prkx
|
protein kinase, X-linked |
| chr7_+_31608828 | 0.31 |
ENSDART00000138491
|
mybpc3
|
myosin binding protein C, cardiac |
| chr23_-_16755682 | 0.31 |
ENSDART00000020810
|
sdcbp2
|
syndecan binding protein (syntenin) 2 |
| chr11_-_34310102 | 0.31 |
ENSDART00000077883
|
pfkfb4a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4a |
| chr22_+_9841541 | 0.31 |
ENSDART00000141085
|
si:dkey-253d23.4
|
si:dkey-253d23.4 |
| chr1_-_37475807 | 0.31 |
ENSDART00000020409
|
hand2
|
heart and neural crest derivatives expressed 2 |
| chr10_+_29964412 | 0.31 |
ENSDART00000117710
|
SNORD14
|
Small nucleolar RNA SNORD14 |
| chr5_+_61678124 | 0.31 |
ENSDART00000074117
|
aspa
|
aspartoacylase |
| chr3_+_19471228 | 0.30 |
ENSDART00000025358
|
itgb3a
|
integrin beta 3a |
| chr4_-_6443699 | 0.30 |
|
|
|
| chr13_+_44720106 | 0.30 |
ENSDART00000017770
|
zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr1_-_11163282 | 0.30 |
ENSDART00000033361
|
ttyh3b
|
tweety family member 3b |
| chr16_+_26566014 | 0.30 |
ENSDART00000041787
|
trim35-28
|
tripartite motif containing 35-28 |
| chr4_+_75262389 | 0.30 |
ENSDART00000174100
|
CABZ01015815.2
|
ENSDARG00000105672 |
| chr7_+_31608878 | 0.30 |
ENSDART00000099789
|
mybpc3
|
myosin binding protein C, cardiac |
| chr11_+_7570326 | 0.30 |
ENSDART00000091550
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| KN150703v1_-_32804 | 0.30 |
|
|
|
| chr2_+_33206349 | 0.29 |
ENSDART00000145588
|
rnf220a
|
ring finger protein 220a |
| chr19_-_10737772 | 0.29 |
ENSDART00000081379
|
olah
|
oleoyl-ACP hydrolase |
| chr8_-_14054145 | 0.29 |
ENSDART00000137857
|
si:ch211-229n2.7
|
si:ch211-229n2.7 |
| chr4_-_71367238 | 0.29 |
ENSDART00000165327
|
CABZ01021450.2
|
ENSDARG00000100390 |
| chr2_-_30071872 | 0.29 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr7_-_60525749 | 0.29 |
ENSDART00000136999
|
pcxb
|
pyruvate carboxylase b |
| chr7_-_73631286 | 0.29 |
ENSDART00000129254
|
zgc:173552
|
zgc:173552 |
| chr11_-_18068313 | 0.29 |
|
|
|
| chr18_+_31094840 | 0.29 |
ENSDART00000023539
|
cyba
|
cytochrome b-245, alpha polypeptide |
| chr11_-_22450525 | 0.29 |
|
|
|
| chr19_-_3880722 | 0.29 |
ENSDART00000169639
|
ENSDARG00000101034
|
ENSDARG00000101034 |
| chr19_+_5501487 | 0.29 |
ENSDART00000132874
|
eif1b
|
eukaryotic translation initiation factor 1B |
| chr2_-_57133471 | 0.28 |
|
|
|
| chr18_-_40518772 | 0.28 |
ENSDART00000021372
|
chrna5
|
cholinergic receptor, nicotinic, alpha 5 |
| chr8_-_49315811 | 0.28 |
ENSDART00000147020
|
prickle3
|
prickle homolog 3 |
| chr11_-_42933969 | 0.28 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr9_-_29056763 | 0.28 |
ENSDART00000060321
|
ENSDARG00000056704
|
ENSDARG00000056704 |
| chr6_+_247024 | 0.28 |
ENSDART00000123769
|
rps19bp1
|
ribosomal protein S19 binding protein 1 |
| chr5_+_56667388 | 0.28 |
ENSDART00000012586
|
slc31a1
|
solute carrier family 31 (copper transporter), member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.2 | GO:0001579 | medium-chain fatty acid transport(GO:0001579) |
| 0.2 | 0.9 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.2 | 0.8 | GO:0019372 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.2 | 0.2 | GO:0071733 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.2 | 0.5 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.2 | 0.6 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.1 | 0.4 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.1 | 0.6 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.4 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.1 | 0.4 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.1 | 0.4 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.1 | 0.4 | GO:0000388 | spliceosome conformational change to release U4 (or U4atac) and U1 (or U11)(GO:0000388) |
| 0.1 | 0.4 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.1 | 0.4 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 0.1 | 0.7 | GO:0071169 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.1 | 0.3 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.1 | 0.8 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.1 | 0.9 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 0.3 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.1 | 0.4 | GO:0071869 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.1 | 0.5 | GO:0031646 | positive regulation of neurological system process(GO:0031646) |
| 0.1 | 0.7 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.1 | 0.3 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.1 | 0.4 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 0.3 | GO:1905178 | regulation of cardiac muscle tissue regeneration(GO:1905178) |
| 0.1 | 0.3 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.1 | 0.2 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.1 | 0.3 | GO:0010939 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) |
| 0.1 | 0.3 | GO:1903726 | negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.1 | 0.2 | GO:0097702 | response to oscillatory fluid shear stress(GO:0097702) cellular response to oscillatory fluid shear stress(GO:0097704) |
| 0.1 | 1.0 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.6 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.1 | 0.4 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 0.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 0.4 | GO:0042539 | hypotonic response(GO:0006971) response to salt stress(GO:0009651) hypotonic salinity response(GO:0042539) |
| 0.1 | 0.6 | GO:0060956 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.1 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.1 | 0.2 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.1 | 0.2 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 0.2 | GO:0006407 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.1 | 0.2 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.1 | 0.4 | GO:0021781 | oligodendrocyte cell fate commitment(GO:0021779) glial cell fate commitment(GO:0021781) |
| 0.1 | 0.3 | GO:0046471 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.1 | 0.3 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.4 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.2 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.1 | GO:2000253 | negative regulation of lipid transport(GO:0032369) endocrine process(GO:0050886) positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.3 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.3 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.7 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.3 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.2 | GO:1900024 | regulation of substrate adhesion-dependent cell spreading(GO:1900024) |
| 0.0 | 0.4 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.0 | 0.2 | GO:0070861 | regulation of COPII vesicle coating(GO:0003400) regulation of protein exit from endoplasmic reticulum(GO:0070861) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.0 | 0.1 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.0 | 0.9 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.6 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 0.3 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.3 | GO:0045453 | bone resorption(GO:0045453) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.0 | 0.2 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 1.8 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.0 | 0.5 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.0 | 0.1 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) |
| 0.0 | 0.8 | GO:0043588 | skin development(GO:0043588) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.7 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0006683 | galactosylceramide catabolic process(GO:0006683) galactolipid catabolic process(GO:0019376) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.2 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.0 | 1.5 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.2 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.1 | GO:0046416 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.0 | 0.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.2 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.2 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.0 | 0.1 | GO:0070544 | negative regulation of ossification(GO:0030279) histone H3-K4 demethylation(GO:0034720) histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.1 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.0 | 0.2 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.0 | 0.6 | GO:0007632 | visual behavior(GO:0007632) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0097198 | histone H3-K36 trimethylation(GO:0097198) |
| 0.0 | 0.4 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.0 | 1.1 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.4 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.0 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.0 | 0.7 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.0 | 0.2 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.0 | 0.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) cellular response to fatty acid(GO:0071398) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.2 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.3 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.0 | GO:0007638 | mechanosensory behavior(GO:0007638) membrane depolarization during action potential(GO:0086010) |
| 0.0 | 0.7 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0019320 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) hexose catabolic process(GO:0019320) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.2 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.1 | GO:0051567 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.0 | 0.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.2 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.2 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.4 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.2 | GO:0050670 | regulation of mononuclear cell proliferation(GO:0032944) regulation of T cell proliferation(GO:0042129) regulation of lymphocyte proliferation(GO:0050670) regulation of leukocyte proliferation(GO:0070663) |
| 0.0 | 0.2 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.2 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.0 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.0 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.0 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.6 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.2 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.4 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:1904949 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.1 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.7 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.4 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.2 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.1 | 0.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 0.2 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.1 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.7 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.7 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 1.0 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 1.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 1.1 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.5 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.3 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.0 | 1.3 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.0 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.0 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.0 | GO:0044304 | main axon(GO:0044304) |
| 0.0 | 0.5 | GO:0098636 | protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.2 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.6 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 1.6 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 0.5 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.2 | 0.8 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.4 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.4 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.1 | 0.9 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 1.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.3 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 0.9 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.3 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 0.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 0.6 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.1 | 0.7 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.1 | 0.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 1.5 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 1.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.2 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.3 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.5 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.4 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.4 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.8 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.4 | GO:0030546 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.0 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0030623 | U5 snRNA binding(GO:0030623) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.1 | GO:0004336 | galactosylceramidase activity(GO:0004336) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0015038 | peptide disulfide oxidoreductase activity(GO:0015037) glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.3 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.1 | GO:0004937 | alpha-adrenergic receptor activity(GO:0004936) alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.3 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.7 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) PDZ domain binding(GO:0030165) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0071855 | neuropeptide hormone activity(GO:0005184) neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.3 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 0.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.1 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0015149 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.0 | 0.1 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.9 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 1.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.6 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.5 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0008443 | 6-phosphofructokinase activity(GO:0003872) phosphofructokinase activity(GO:0008443) fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 2.3 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.0 | 0.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.0 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.7 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.1 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.2 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.0 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.1 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 1.0 | GO:0020037 | heme binding(GO:0020037) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.7 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.3 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.2 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 0.7 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 0.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.2 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 0.4 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |