Project
DANIO-CODE
Navigation
Downloads
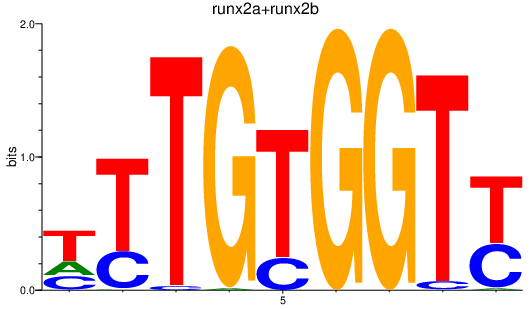
Results for runx2a+runx2b
Z-value: 1.89
Transcription factors associated with runx2a+runx2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
runx2a
|
ENSDARG00000040261 | RUNX family transcription factor 2a |
|
runx2b
|
ENSDARG00000059233 | RUNX family transcription factor 2b |
Activity profile of runx2a+runx2b motif
Sorted Z-values of runx2a+runx2b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of runx2a+runx2b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_24093289 | 8.82 |
ENSDART00000064789
|
txn
|
thioredoxin |
| chr21_-_99353 | 5.71 |
ENSDART00000040422
|
bhmt
|
betaine-homocysteine methyltransferase |
| chr7_-_50827308 | 5.07 |
ENSDART00000121574
|
col4a6
|
collagen, type IV, alpha 6 |
| chr11_+_37639045 | 4.63 |
ENSDART00000111157
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr13_-_39033893 | 4.04 |
ENSDART00000045434
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr24_-_4941954 | 3.97 |
ENSDART00000127597
|
zic1
|
zic family member 1 (odd-paired homolog, Drosophila) |
| chr25_+_5232865 | 3.95 |
|
|
|
| chr7_+_25758469 | 3.86 |
ENSDART00000101126
|
alox12
|
arachidonate 12-lipoxygenase |
| chr23_+_23730717 | 3.83 |
|
|
|
| chr9_-_37940101 | 3.78 |
ENSDART00000087663
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr8_+_23759741 | 3.62 |
ENSDART00000162924
|
SCARNA2
|
Small Cajal body-specific RNA 2 |
| chr23_+_36002332 | 3.58 |
ENSDART00000103139
|
hoxc8a
|
homeobox C8a |
| chr9_-_14533551 | 3.41 |
ENSDART00000056103
|
nrp2b
|
neuropilin 2b |
| chr16_-_9978112 | 3.33 |
ENSDART00000149312
|
ncalda
|
neurocalcin delta a |
| chr5_+_56667388 | 3.32 |
ENSDART00000012586
|
slc31a1
|
solute carrier family 31 (copper transporter), member 1 |
| chr9_-_23307419 | 3.24 |
ENSDART00000020884
|
lypd6
|
LY6/PLAUR domain containing 6 |
| chr25_+_7858886 | 3.23 |
ENSDART00000171904
|
ucmab
|
upper zone of growth plate and cartilage matrix associated b |
| chr1_+_46537108 | 3.23 |
|
|
|
| chr14_+_38445969 | 3.21 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr6_+_56163589 | 3.19 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr23_+_36023748 | 3.16 |
|
|
|
| chr24_+_11194381 | 3.06 |
ENSDART00000143171
|
si:dkey-12l12.1
|
si:dkey-12l12.1 |
| chr6_-_7281412 | 3.03 |
ENSDART00000053776
|
fkbp11
|
FK506 binding protein 11 |
| chr21_+_13063614 | 3.02 |
|
|
|
| chr14_-_23953804 | 3.00 |
ENSDART00000114169
|
bnip1a
|
BCL2/adenovirus E1B interacting protein 1a |
| chr20_+_52653111 | 2.99 |
ENSDART00000057980
|
tsta3
|
tissue specific transplantation antigen P35B |
| chr1_+_1953714 | 2.98 |
ENSDART00000164488
ENSDART00000167050 ENSDART00000122626 ENSDART00000128187 |
mbnl2
|
muscleblind-like splicing regulator 2 |
| chr19_+_31046291 | 2.98 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr11_-_23151247 | 2.95 |
|
|
|
| chr1_+_1541977 | 2.94 |
ENSDART00000048828
|
atp1a1a.4
|
ATPase, Na+/K+ transporting, alpha 1a polypeptide, tandem duplicate 4 |
| chr21_-_20305406 | 2.92 |
ENSDART00000065659
|
rbp4l
|
retinol binding protein 4, like |
| chr21_-_7703717 | 2.89 |
ENSDART00000158852
|
egfl7
|
EGF-like-domain, multiple 7 |
| chr11_+_8119829 | 2.85 |
ENSDART00000011183
|
prkacba
|
protein kinase, cAMP-dependent, catalytic, beta a |
| chr21_-_11539798 | 2.84 |
ENSDART00000144770
|
cast
|
calpastatin |
| chr24_-_21778717 | 2.81 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr2_-_30198789 | 2.80 |
ENSDART00000019149
|
rpl7
|
ribosomal protein L7 |
| chr7_-_11812634 | 2.79 |
ENSDART00000101537
|
mex3b
|
mex-3 RNA binding family member B |
| chr20_-_14158114 | 2.77 |
ENSDART00000009549
|
rhag
|
Rh-associated glycoprotein |
| chr23_-_2084541 | 2.72 |
ENSDART00000091532
|
ndnf
|
neuron-derived neurotrophic factor |
| chr22_+_9841541 | 2.70 |
ENSDART00000141085
|
si:dkey-253d23.4
|
si:dkey-253d23.4 |
| chr9_-_29056763 | 2.70 |
ENSDART00000060321
|
ENSDARG00000056704
|
ENSDARG00000056704 |
| chr24_+_17125429 | 2.69 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr1_-_37475807 | 2.64 |
ENSDART00000020409
|
hand2
|
heart and neural crest derivatives expressed 2 |
| chr4_-_19895115 | 2.64 |
ENSDART00000105967
|
cacna2d1a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1a |
| chr11_-_11591394 | 2.63 |
ENSDART00000142208
|
zgc:110712
|
zgc:110712 |
| chr20_-_35343242 | 2.62 |
ENSDART00000113294
|
fzd3a
|
frizzled class receptor 3a |
| chr12_-_8448482 | 2.62 |
ENSDART00000062855
|
egr2b
|
early growth response 2b |
| chr18_-_44914637 | 2.62 |
ENSDART00000169636
|
si:ch211-71n6.4
|
si:ch211-71n6.4 |
| chr3_+_31490043 | 2.60 |
ENSDART00000076636
|
fzd2
|
frizzled class receptor 2 |
| chr19_+_22478256 | 2.60 |
ENSDART00000100181
|
sall3b
|
spalt-like transcription factor 3b |
| chr20_+_20600211 | 2.55 |
ENSDART00000036124
|
six1b
|
SIX homeobox 1b |
| chr18_+_38307946 | 2.54 |
ENSDART00000134247
|
lmo2
|
LIM domain only 2 (rhombotin-like 1) |
| chr22_-_10430130 | 2.53 |
ENSDART00000064801
|
ogn
|
osteoglycin |
| chr3_-_40159349 | 2.52 |
ENSDART00000055186
|
atp5j2
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F2 |
| chr10_-_25695574 | 2.52 |
ENSDART00000110751
|
tiam1a
|
T-cell lymphoma invasion and metastasis 1a |
| chr19_+_16318256 | 2.51 |
ENSDART00000137189
|
ptprua
|
protein tyrosine phosphatase, receptor type, U, a |
| chr15_-_42250788 | 2.49 |
ENSDART00000015843
|
pax3b
|
paired box 3b |
| chr7_+_25587183 | 2.47 |
ENSDART00000148780
|
mtmr1a
|
myotubularin related protein 1a |
| chr16_-_29593569 | 2.46 |
ENSDART00000150028
|
onecutl
|
one cut domain, family member, like |
| chr7_-_48392300 | 2.46 |
|
|
|
| chr14_+_6117282 | 2.43 |
ENSDART00000051556
|
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr13_+_22545318 | 2.41 |
ENSDART00000143312
|
zgc:193505
|
zgc:193505 |
| chr2_+_21067708 | 2.41 |
ENSDART00000148400
ENSDART00000021168 |
rxrga
|
retinoid x receptor, gamma a |
| chr13_-_33574216 | 2.39 |
ENSDART00000065435
|
cst3
|
cystatin C (amyloid angiopathy and cerebral hemorrhage) |
| chr8_+_21321765 | 2.39 |
ENSDART00000131691
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr16_-_42063851 | 2.39 |
ENSDART00000045403
|
etv2
|
ets variant 2 |
| chr23_+_28656263 | 2.38 |
ENSDART00000020296
|
nadl1.2
|
neural adhesion molecule L1.2 |
| chr2_+_24649130 | 2.34 |
ENSDART00000078972
|
fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr25_+_30746445 | 2.32 |
ENSDART00000156916
|
lsp1
|
lymphocyte-specific protein 1 |
| chr2_-_40170303 | 2.32 |
ENSDART00000165602
|
epha4a
|
eph receptor A4a |
| chr9_-_42894582 | 2.28 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr19_-_22904687 | 2.28 |
ENSDART00000141503
|
pleca
|
plectin a |
| chr5_+_34022151 | 2.26 |
ENSDART00000141338
|
enc1
|
ectodermal-neural cortex 1 |
| chr11_-_38940966 | 2.26 |
ENSDART00000105133
|
wnt4a
|
wingless-type MMTV integration site family, member 4a |
| chr10_-_43924675 | 2.25 |
ENSDART00000052307
|
arrdc3b
|
arrestin domain containing 3b |
| chr20_+_54289892 | 2.23 |
ENSDART00000060444
|
rps29
|
ribosomal protein S29 |
| chr19_+_17461581 | 2.23 |
ENSDART00000159838
|
lye
|
lymphocyte antigen-6, epidermis |
| chr2_-_30675594 | 2.23 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr18_+_12207888 | 2.22 |
|
|
|
| chr6_+_45917081 | 2.21 |
ENSDART00000149450
ENSDART00000149642 |
h6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr12_-_26760324 | 2.20 |
ENSDART00000047724
|
zeb1b
|
zinc finger E-box binding homeobox 1b |
| chr15_+_19902697 | 2.20 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr13_-_42986821 | 2.19 |
ENSDART00000164439
|
si:ch211-106f21.1
|
si:ch211-106f21.1 |
| chr15_-_10345337 | 2.16 |
|
|
|
| chr19_+_14103214 | 2.16 |
|
|
|
| chr1_-_24486146 | 2.15 |
ENSDART00000144711
|
tmem154
|
transmembrane protein 154 |
| chr11_-_18004437 | 2.14 |
ENSDART00000125800
|
CABZ01112210.1
|
ENSDARG00000088766 |
| chr4_+_15870605 | 2.13 |
ENSDART00000178674
|
BX004864.1
|
ENSDARG00000107641 |
| chr3_+_19471228 | 2.13 |
ENSDART00000025358
|
itgb3a
|
integrin beta 3a |
| chr8_-_4903896 | 2.12 |
ENSDART00000004986
|
igsf9b
|
immunoglobulin superfamily, member 9b |
| chr21_+_7844259 | 2.12 |
ENSDART00000027268
|
otpa
|
orthopedia homeobox a |
| chr15_-_23441268 | 2.08 |
ENSDART00000078570
|
c1qtnf5
|
C1q and TNF related 5 |
| chr1_+_49894978 | 2.08 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr22_+_18927612 | 2.08 |
ENSDART00000136390
|
hcn2b
|
hyperpolarization activated cyclic nucleotide-gated potassium channel 2b |
| chr23_-_15381030 | 2.07 |
|
|
|
| chr16_+_33041823 | 2.07 |
ENSDART00000170157
|
prss35
|
protease, serine, 35 |
| chr3_-_57691670 | 2.05 |
ENSDART00000156522
|
cant1a
|
calcium activated nucleotidase 1a |
| chr15_-_18066180 | 2.03 |
ENSDART00000154524
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr1_-_44886649 | 2.03 |
|
|
|
| chr21_-_26459113 | 2.02 |
ENSDART00000157255
|
cd248b
|
CD248 molecule, endosialin b |
| chr23_-_20837809 | 2.01 |
ENSDART00000089750
|
znf362b
|
zinc finger protein 362b |
| chr22_+_13862110 | 1.99 |
ENSDART00000105711
|
sh3bp4a
|
SH3-domain binding protein 4a |
| chr8_-_41266057 | 1.99 |
ENSDART00000108518
|
rnf10
|
ring finger protein 10 |
| chr2_-_9950224 | 1.97 |
ENSDART00000146407
|
si:ch1073-170o4.1
|
si:ch1073-170o4.1 |
| chr20_-_35343057 | 1.97 |
ENSDART00000113294
|
fzd3a
|
frizzled class receptor 3a |
| KN150256v1_-_9467 | 1.96 |
|
|
|
| chr11_+_5467173 | 1.95 |
|
|
|
| chr3_-_39031305 | 1.95 |
ENSDART00000022393
|
si:dkeyp-57f11.2
|
si:dkeyp-57f11.2 |
| chr9_+_36051713 | 1.93 |
ENSDART00000134447
|
rcan1a
|
regulator of calcineurin 1a |
| chr16_-_16796313 | 1.91 |
|
|
|
| chr24_+_37834148 | 1.88 |
|
|
|
| chr25_-_22890657 | 1.86 |
|
|
|
| chr2_+_54060488 | 1.86 |
ENSDART00000167239
|
CABZ01050249.1
|
ENSDARG00000099376 |
| chr13_-_36996246 | 1.85 |
ENSDART00000133242
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr8_+_7297981 | 1.85 |
ENSDART00000170975
|
BX469901.2
|
ENSDARG00000103232 |
| chr14_+_34174018 | 1.84 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr16_+_38327295 | 1.79 |
ENSDART00000085143
|
bnipl
|
BCL2/adenovirus E1B 19kD interacting protein, like |
| chr17_-_9869142 | 1.79 |
ENSDART00000008355
|
cfl2
|
cofilin 2 (muscle) |
| chr4_-_11604184 | 1.76 |
ENSDART00000170136
|
net1
|
neuroepithelial cell transforming 1 |
| chr15_+_30442890 | 1.76 |
ENSDART00000048847
|
nos2b
|
nitric oxide synthase 2b, inducible |
| chr14_+_4489377 | 1.75 |
ENSDART00000041468
|
ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr16_-_24280209 | 1.73 |
ENSDART00000048599
|
rps19
|
ribosomal protein S19 |
| chr17_-_2513630 | 1.73 |
ENSDART00000135374
|
ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr5_+_26252925 | 1.73 |
|
|
|
| chr24_+_19374200 | 1.72 |
ENSDART00000056081
ENSDART00000027022 |
sulf1
|
sulfatase 1 |
| chr6_-_7281346 | 1.71 |
ENSDART00000133096
|
fkbp11
|
FK506 binding protein 11 |
| chr10_+_10393377 | 1.71 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr4_-_16417703 | 1.71 |
ENSDART00000013085
|
dcn
|
decorin |
| chr16_+_42067930 | 1.71 |
ENSDART00000102789
|
fli1b
|
Fli-1 proto-oncogene, ETS transcription factor b |
| chr23_+_34079219 | 1.67 |
ENSDART00000132668
|
si:ch211-207e14.4
|
si:ch211-207e14.4 |
| chr18_+_6692992 | 1.67 |
|
|
|
| chr5_-_31845371 | 1.66 |
ENSDART00000157295
ENSDART00000159522 |
ncs1a
|
neuronal calcium sensor 1a |
| chr2_-_30071872 | 1.63 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr7_-_33558771 | 1.63 |
ENSDART00000074729
|
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr20_-_20634087 | 1.62 |
ENSDART00000125039
|
six6b
|
SIX homeobox 6b |
| chr4_-_19895001 | 1.62 |
ENSDART00000014440
|
cacna2d1a
|
calcium channel, voltage-dependent, alpha 2/delta subunit 1a |
| chr1_-_38189932 | 1.61 |
ENSDART00000136102
|
vegfc
|
vascular endothelial growth factor c |
| chr7_-_18346474 | 1.59 |
|
|
|
| chr22_+_5657904 | 1.59 |
ENSDART00000138102
|
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr7_+_25649559 | 1.59 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr13_-_4204592 | 1.58 |
ENSDART00000122406
|
znf318
|
zinc finger protein 318 |
| chr9_-_47041677 | 1.57 |
ENSDART00000054137
|
igfbp5b
|
insulin-like growth factor binding protein 5b |
| chr11_-_18004011 | 1.57 |
ENSDART00000125800
|
CABZ01112210.1
|
ENSDARG00000088766 |
| chr17_+_48999061 | 1.56 |
ENSDART00000177390
|
tiam2a
|
T-cell lymphoma invasion and metastasis 2a |
| chr4_+_8531280 | 1.55 |
ENSDART00000162065
|
wnt5b
|
wingless-type MMTV integration site family, member 5b |
| chr15_-_35394535 | 1.54 |
ENSDART00000144153
|
mff
|
mitochondrial fission factor |
| chr7_+_59773957 | 1.54 |
ENSDART00000051524
|
etnppl
|
ethanolamine-phosphate phospho-lyase |
| chr22_+_17091411 | 1.53 |
ENSDART00000170076
|
frem1b
|
Fras1 related extracellular matrix 1b |
| chr13_+_1484997 | 1.53 |
|
|
|
| chr23_-_29138952 | 1.52 |
ENSDART00000002812
|
casz1
|
castor zinc finger 1 |
| chr1_-_19001718 | 1.52 |
ENSDART00000143303
|
BX323590.2
|
ENSDARG00000095388 |
| chr14_-_14260824 | 1.51 |
ENSDART00000159056
|
si:dkey-27i16.2
|
si:dkey-27i16.2 |
| chr6_-_24003717 | 1.50 |
ENSDART00000164915
|
scinla
|
scinderin like a |
| chr7_-_40713381 | 1.49 |
ENSDART00000031700
|
en2a
|
engrailed homeobox 2a |
| chr23_+_22051774 | 1.49 |
ENSDART00000145172
|
eif4g3b
|
eukaryotic translation initiation factor 4 gamma, 3b |
| chr6_-_12079553 | 1.48 |
ENSDART00000157058
|
si:dkey-276j7.1
|
si:dkey-276j7.1 |
| chr17_+_16557246 | 1.48 |
ENSDART00000015729
ENSDART00000136874 |
foxn3
|
forkhead box N3 |
| chr15_-_24934442 | 1.46 |
ENSDART00000127047
|
tusc5a
|
tumor suppressor candidate 5a |
| chr19_+_3925188 | 1.46 |
|
|
|
| chr2_-_44429891 | 1.46 |
ENSDART00000163040
ENSDART00000056372 ENSDART00000109251 ENSDART00000166923 ENSDART00000132682 |
mpz
|
myelin protein zero |
| chr20_+_18803167 | 1.46 |
ENSDART00000019476
|
eif5
|
eukaryotic translation initiation factor 5 |
| chr4_-_25080470 | 1.46 |
ENSDART00000179640
|
gata3
|
GATA binding protein 3 |
| chr8_-_38308535 | 1.45 |
|
|
|
| chr3_+_55779906 | 1.45 |
|
|
|
| chr16_-_22947908 | 1.45 |
ENSDART00000133910
|
shc1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr22_-_14247636 | 1.44 |
|
|
|
| chr8_-_23744125 | 1.42 |
ENSDART00000141871
|
INAVA
|
innate immunity activator |
| chr9_-_306569 | 1.41 |
ENSDART00000166059
|
si:ch211-166e11.5
|
si:ch211-166e11.5 |
| chr2_+_23038657 | 1.41 |
ENSDART00000089012
|
kif1ab
|
kinesin family member 1Ab |
| chr9_-_695593 | 1.41 |
ENSDART00000115030
|
dip2a
|
disco-interacting protein 2 homolog A |
| chr6_+_48156180 | 1.40 |
ENSDART00000019706
|
phc2b
|
polyhomeotic homolog 2b (Drosophila) |
| chr13_-_10128903 | 1.39 |
ENSDART00000080808
|
six3a
|
SIX homeobox 3a |
| chr13_+_10100146 | 1.38 |
ENSDART00000080805
|
six2a
|
SIX homeobox 2a |
| chr20_-_3970778 | 1.38 |
ENSDART00000178724
ENSDART00000178565 |
TRIM67
|
tripartite motif containing 67 |
| chr18_+_22120282 | 1.38 |
ENSDART00000147230
|
zgc:158868
|
zgc:158868 |
| chr21_-_2143830 | 1.37 |
|
|
|
| chr14_+_6639864 | 1.36 |
ENSDART00000061001
|
gnb2l1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr22_-_14247765 | 1.35 |
|
|
|
| chr5_-_31845561 | 1.35 |
ENSDART00000157295
ENSDART00000159522 |
ncs1a
|
neuronal calcium sensor 1a |
| chr1_-_57842604 | 1.33 |
|
|
|
| chr23_-_6707246 | 1.32 |
ENSDART00000023793
|
mipb
|
major intrinsic protein of lens fiber b |
| chr16_+_11833310 | 1.31 |
ENSDART00000060266
|
ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1 |
| chr13_-_10128836 | 1.31 |
ENSDART00000080808
|
six3a
|
SIX homeobox 3a |
| chr21_-_3548719 | 1.31 |
ENSDART00000137844
|
atp8b1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr7_+_55883471 | 1.31 |
ENSDART00000073594
|
ankrd11
|
ankyrin repeat domain 11 |
| chr1_+_14451504 | 1.31 |
ENSDART00000111475
|
pcdh7a
|
protocadherin 7a |
| chr16_+_25425398 | 1.28 |
|
|
|
| chr18_-_41384938 | 1.28 |
ENSDART00000098673
|
ptx3a
|
pentraxin 3, long a |
| chr24_+_2322328 | 1.28 |
ENSDART00000132530
|
si:ch211-266d19.4
|
si:ch211-266d19.4 |
| chr5_+_23093316 | 1.28 |
ENSDART00000006983
|
kat5a
|
K(lysine) acetyltransferase 5a |
| chr4_+_14361655 | 1.27 |
ENSDART00000007103
|
nuak1a
|
NUAK family, SNF1-like kinase, 1a |
| chr5_-_28025315 | 1.27 |
ENSDART00000131729
|
tnc
|
tenascin C |
| chr7_+_34394526 | 1.27 |
|
|
|
| chr10_-_7954888 | 1.27 |
ENSDART00000139661
|
slc35e4
|
solute carrier family 35, member E4 |
| chr9_-_22394674 | 1.27 |
ENSDART00000101869
|
crygm2d12
|
crystallin, gamma M2d12 |
| chr24_+_25326286 | 1.26 |
ENSDART00000066625
|
smpx
|
small muscle protein, X-linked |
| chr23_-_24240728 | 1.26 |
ENSDART00000113598
|
arhgef19
|
Rho guanine nucleotide exchange factor (GEF) 19 |
| chr1_-_41281418 | 1.25 |
ENSDART00000014678
|
adra1d
|
adrenoceptor alpha 1D |
| chr1_-_57329034 | 1.25 |
ENSDART00000160970
|
caspb
|
caspase b |
| chr11_+_23694891 | 1.25 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.6 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 1.2 | 7.3 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 1.1 | 3.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 1.0 | 3.0 | GO:0097702 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) response to oscillatory fluid shear stress(GO:0097702) cellular response to oscillatory fluid shear stress(GO:0097704) |
| 1.0 | 3.9 | GO:0051122 | lipoxygenase pathway(GO:0019372) linoleic acid metabolic process(GO:0043651) hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.9 | 2.7 | GO:0006929 | substrate-dependent cell migration(GO:0006929) |
| 0.9 | 2.6 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.8 | 3.4 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.8 | 7.5 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.8 | 4.0 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.7 | 2.2 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.7 | 2.2 | GO:1904105 | positive regulation of convergent extension involved in gastrulation(GO:1904105) |
| 0.7 | 3.3 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.6 | 1.9 | GO:0045887 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) positive regulation of neuromuscular junction development(GO:1904398) |
| 0.6 | 2.5 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.6 | 1.8 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.6 | 1.8 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.6 | 2.8 | GO:0031646 | positive regulation of neurological system process(GO:0031646) |
| 0.5 | 1.6 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.5 | 1.6 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) regulation of protein binding(GO:0043393) |
| 0.5 | 4.6 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.5 | 2.5 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.5 | 3.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.4 | 1.8 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.4 | 2.6 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.4 | 2.5 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.4 | 1.3 | GO:0003321 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.4 | 3.2 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.4 | 1.2 | GO:1902024 | serine transport(GO:0032329) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.4 | 1.6 | GO:0033499 | galactose catabolic process(GO:0019388) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.4 | 3.4 | GO:0071678 | olfactory bulb axon guidance(GO:0071678) |
| 0.4 | 3.2 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.3 | 3.8 | GO:0003348 | cardiac endothelial cell differentiation(GO:0003348) endocardial cell differentiation(GO:0060956) |
| 0.3 | 1.0 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.3 | 2.3 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.3 | 2.8 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.3 | 1.3 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.3 | 1.2 | GO:0035776 | pronephric proximal tubule development(GO:0035776) |
| 0.3 | 2.4 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.3 | 1.8 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.3 | 0.9 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.3 | 1.8 | GO:1900024 | regulation of substrate adhesion-dependent cell spreading(GO:1900024) |
| 0.3 | 2.6 | GO:0048937 | lateral line nerve glial cell differentiation(GO:0048895) myelination of lateral line nerve axons(GO:0048897) posterior lateral line nerve glial cell differentiation(GO:0048931) myelination of posterior lateral line nerve axons(GO:0048932) lateral line nerve glial cell development(GO:0048937) lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048938) posterior lateral line nerve glial cell development(GO:0048941) posterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048942) |
| 0.3 | 1.5 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 0.3 | 0.9 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.3 | 1.1 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 0.3 | 5.1 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.3 | 4.7 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.3 | 1.6 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.3 | 1.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.3 | 4.7 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.3 | 2.6 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.3 | 2.6 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.3 | 1.3 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) cellular response to fatty acid(GO:0071398) |
| 0.2 | 2.7 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.2 | 2.2 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.2 | 1.6 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.2 | 0.8 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 0.8 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.2 | 3.2 | GO:0014036 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.2 | 2.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.2 | 2.2 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.2 | 5.2 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.2 | 2.1 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.2 | 0.7 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 | 3.7 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.2 | 0.7 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) response to cAMP(GO:0051591) cellular response to cAMP(GO:0071320) negative regulation of synaptic vesicle transport(GO:1902804) negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.2 | 0.5 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 2.9 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.1 | 2.8 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.1 | 2.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 1.0 | GO:0006735 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 3.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 2.4 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 4.4 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.1 | 1.0 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 1.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 0.8 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.1 | 1.6 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.1 | 1.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 5.3 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.1 | 0.8 | GO:0007584 | response to nutrient(GO:0007584) |
| 0.1 | 0.6 | GO:0009180 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.1 | 1.2 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.1 | 0.4 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.1 | 0.9 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.9 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.1 | 0.5 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 0.9 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.0 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.9 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 2.9 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 1.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 2.9 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.1 | 0.3 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.3 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 3.2 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 1.3 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 0.2 | GO:0051029 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.1 | 0.9 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.1 | 0.5 | GO:0021602 | cranial nerve morphogenesis(GO:0021602) |
| 0.1 | 1.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.1 | 0.4 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 1.3 | GO:0001843 | neural tube closure(GO:0001843) tube closure(GO:0060606) |
| 0.0 | 4.7 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 1.2 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.0 | 0.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 3.4 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 1.7 | GO:0001837 | epithelial to mesenchymal transition(GO:0001837) |
| 0.0 | 0.3 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.0 | 1.3 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 0.8 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.7 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.5 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.7 | GO:0048488 | synaptic vesicle endocytosis(GO:0048488) |
| 0.0 | 1.3 | GO:0030166 | proteoglycan biosynthetic process(GO:0030166) |
| 0.0 | 3.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.8 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.5 | GO:0006813 | potassium ion transport(GO:0006813) |
| 0.0 | 0.6 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.6 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 0.1 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.4 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 1.1 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 1.9 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.9 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.6 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.3 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.2 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 1.0 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.6 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.4 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.9 | 4.3 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.4 | 3.0 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.3 | 1.0 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.3 | 1.3 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.3 | 1.3 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 1.0 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 3.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.2 | 1.3 | GO:0016234 | inclusion body(GO:0016234) |
| 0.2 | 2.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.2 | 1.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.2 | 2.1 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 1.0 | GO:0097651 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 0.9 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 1.5 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 3.0 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 2.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 5.0 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 4.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 1.3 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.1 | 0.9 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.2 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.1 | 0.3 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.1 | 2.8 | GO:0022626 | cytosolic large ribosomal subunit(GO:0022625) cytosolic ribosome(GO:0022626) |
| 0.1 | 9.1 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.9 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.0 | 3.0 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 1.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 22.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.5 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 1.1 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 2.1 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 4.0 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.3 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 3.8 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 2.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.4 | GO:0044420 | extracellular matrix component(GO:0044420) |
| 0.0 | 3.9 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.0 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 1.1 | GO:0012506 | vesicle membrane(GO:0012506) |
| 0.0 | 0.5 | GO:0005643 | nuclear pore(GO:0005643) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.6 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 1.1 | 3.2 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.7 | 2.2 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.7 | 2.0 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.7 | 3.3 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.6 | 1.8 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.6 | 2.9 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.5 | 3.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.5 | 2.4 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.4 | 4.0 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.4 | 2.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.4 | 4.9 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.4 | 1.7 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.4 | 1.7 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.4 | 2.4 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.4 | 3.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.4 | 7.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.4 | 3.8 | GO:0005113 | patched binding(GO:0005113) |
| 0.4 | 1.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.4 | 4.7 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.4 | 3.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.3 | 2.8 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.3 | 8.8 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.3 | 1.3 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) alpha1-adrenergic receptor activity(GO:0004937) |
| 0.3 | 1.8 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.3 | 1.6 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.3 | 3.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.3 | 1.6 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.3 | 1.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 2.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 0.9 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.2 | 0.7 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.2 | 0.6 | GO:0000829 | inositol heptakisphosphate kinase activity(GO:0000829) diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.2 | 1.3 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.2 | 2.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.2 | 2.1 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.2 | 1.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.2 | 1.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 1.5 | GO:0016838 | carbon-oxygen lyase activity, acting on phosphates(GO:0016838) |
| 0.2 | 2.7 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.2 | 0.7 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 3.9 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 9.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 4.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 1.6 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 1.6 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 1.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 1.2 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 1.0 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.8 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.1 | 4.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.1 | 1.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 3.1 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.1 | 0.5 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.9 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.9 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 0.9 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 0.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 2.9 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 3.8 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 2.4 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.1 | 0.5 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 3.2 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.1 | GO:0005112 | Notch binding(GO:0005112) PDZ domain binding(GO:0030165) |
| 0.1 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.1 | 2.7 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 3.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 0.6 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.3 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 5.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 4.8 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.5 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 6.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.7 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 1.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.7 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.3 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.0 | 0.4 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 1.3 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 0.3 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.2 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 2.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.5 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 31.7 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 0.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.7 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 9.4 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.1 | GO:0019956 | chemokine binding(GO:0019956) C-C chemokine binding(GO:0019957) |
| 0.0 | 1.5 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 2.3 | GO:0042393 | histone binding(GO:0042393) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.4 | 8.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.3 | 7.4 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 4.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 2.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 1.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 2.8 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.1 | 1.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.6 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.1 | 3.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 1.0 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.1 | 0.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.4 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.1 | 5.1 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 2.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.5 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 1.0 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.8 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.0 | 1.0 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.5 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 2.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 8.8 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.4 | 1.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.3 | 2.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.3 | 1.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.3 | 2.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 1.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 2.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 2.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 6.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.2 | 3.2 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.2 | 5.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.7 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 1.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 4.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 0.7 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.1 | 1.0 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 1.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.5 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 2.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 2.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 2.6 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 2.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 1.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 0.3 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.6 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.5 | REACTOME IL 3 5 AND GM CSF SIGNALING | Genes involved in Interleukin-3, 5 and GM-CSF signaling |
| 0.0 | 0.9 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 1.5 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 1.6 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |