Project
DANIO-CODE
Navigation
Downloads
Results for runx3
Z-value: 0.55
Transcription factors associated with runx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
runx3
|
ENSDARG00000052826 | RUNX family transcription factor 3 |
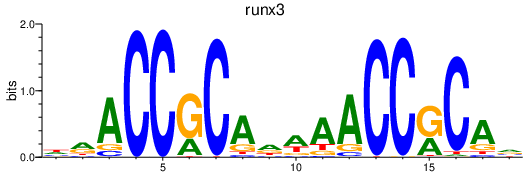
Activity profile of runx3 motif
Sorted Z-values of runx3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of runx3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_50066633 | 2.11 |
|
|
|
| chr6_-_9346394 | 1.42 |
ENSDART00000144335
|
cyp27c1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr5_+_26829686 | 1.15 |
ENSDART00000087894
|
zgc:153409
|
zgc:153409 |
| chr23_+_358044 | 1.14 |
ENSDART00000114000
|
zgc:101663
|
zgc:101663 |
| chr14_-_3264798 | 1.11 |
ENSDART00000171601
|
im:7150988
|
im:7150988 |
| chr8_+_17148864 | 1.04 |
ENSDART00000140531
|
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr15_-_20532537 | 0.96 |
ENSDART00000018514
|
dlc
|
deltaC |
| chr9_+_37331936 | 0.92 |
ENSDART00000024555
|
gli2a
|
GLI family zinc finger 2a |
| chr5_+_68889557 | 0.89 |
ENSDART00000160171
|
CT978957.3
|
ENSDARG00000103717 |
| chr9_-_20562293 | 0.75 |
ENSDART00000113418
|
igsf3
|
immunoglobulin superfamily, member 3 |
| chr11_-_40255633 | 0.71 |
ENSDART00000172819
|
miip
|
migration and invasion inhibitory protein |
| chr16_+_23516127 | 0.71 |
ENSDART00000004679
|
icn
|
ictacalcin |
| chr19_-_5464238 | 0.69 |
ENSDART00000105806
|
cyt1
|
type I cytokeratin, enveloping layer |
| chr14_-_16171165 | 0.68 |
ENSDART00000089021
|
canx
|
calnexin |
| chr4_-_71353374 | 0.59 |
|
|
|
| chr19_-_24859287 | 0.59 |
ENSDART00000163763
|
thbs3b
|
thrombospondin 3b |
| chr16_+_28819826 | 0.58 |
ENSDART00000103340
|
s100v1
|
S100 calcium binding protein V1 |
| chr12_+_18622682 | 0.57 |
ENSDART00000153456
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr4_-_689860 | 0.55 |
|
|
|
| chr8_-_18868986 | 0.54 |
ENSDART00000079840
|
rorca
|
RAR-related orphan receptor C a |
| chr2_-_56224858 | 0.52 |
ENSDART00000060745
|
uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr3_-_25239180 | 0.48 |
ENSDART00000055491
|
smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr22_+_10728099 | 0.47 |
ENSDART00000145551
|
tmprss9
|
transmembrane protease, serine 9 |
| KN150099v1_-_34647 | 0.46 |
ENSDART00000157438
|
nudt12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr11_-_17620732 | 0.46 |
ENSDART00000154627
|
eogt
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr12_+_28795950 | 0.44 |
ENSDART00000076572
|
rnf40
|
ring finger protein 40 |
| chr3_-_45237479 | 0.43 |
ENSDART00000160923
|
si:ch73-167f10.1
|
si:ch73-167f10.1 |
| chr22_+_11490729 | 0.42 |
ENSDART00000063147
|
sgsh
|
N-sulfoglucosamine sulfohydrolase (sulfamidase) |
| chr5_-_33611896 | 0.41 |
|
|
|
| chr1_+_49894685 | 0.41 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr14_-_223462 | 0.39 |
|
|
|
| chr4_+_689631 | 0.38 |
|
|
|
| chr13_-_31516518 | 0.38 |
ENSDART00000172375
ENSDART00000125987 ENSDART00000143903 ENSDART00000158719 |
six4a
|
SIX homeobox 4a |
| chr20_-_43448899 | 0.36 |
ENSDART00000152888
|
mllt4a
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 4a |
| chr8_+_17148832 | 0.35 |
ENSDART00000140531
|
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr16_+_41110126 | 0.35 |
ENSDART00000141130
|
scap
|
SREBF chaperone |
| chr18_+_35198982 | 0.34 |
ENSDART00000127379
|
cfap45
|
cilia and flagella associated protein 45 |
| KN149797v1_-_53588 | 0.34 |
|
|
|
| chr6_+_54146669 | 0.32 |
ENSDART00000061735
|
nudt3b
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3b |
| chr9_-_23936295 | 0.29 |
ENSDART00000165907
|
ndufa10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10 |
| chr21_-_43020159 | 0.27 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr3_-_25238896 | 0.26 |
ENSDART00000055491
|
smurf2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr18_-_50527399 | 0.26 |
ENSDART00000033591
|
cd276
|
CD276 molecule |
| chr24_-_14068113 | 0.26 |
ENSDART00000130825
|
xkr9
|
XK, Kell blood group complex subunit-related family, member 9 |
| chr2_-_7375138 | 0.26 |
ENSDART00000146434
|
zgc:153115
|
zgc:153115 |
| chr12_-_151380 | 0.23 |
|
|
|
| chr12_+_28795771 | 0.23 |
ENSDART00000076342
|
rnf40
|
ring finger protein 40 |
| chr20_-_34767188 | 0.23 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr10_+_45301543 | 0.22 |
ENSDART00000172621
|
ogdhb
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase b (lipoamide) |
| chr8_-_17148743 | 0.21 |
ENSDART00000025803
|
pola2
|
polymerase (DNA directed), alpha 2 |
| chr3_-_3589844 | 0.21 |
ENSDART00000140482
|
ENSDARG00000079723
|
ENSDARG00000079723 |
| chr12_+_46867586 | 0.21 |
ENSDART00000008312
|
fam53b
|
family with sequence similarity 53, member B |
| chr17_-_37247505 | 0.21 |
ENSDART00000108514
|
asxl2
|
additional sex combs like transcriptional regulator 2 |
| chr13_+_30190977 | 0.20 |
|
|
|
| chr7_+_25587183 | 0.19 |
ENSDART00000148780
|
mtmr1a
|
myotubularin related protein 1a |
| chr13_-_31516399 | 0.19 |
ENSDART00000172375
ENSDART00000125987 ENSDART00000143903 ENSDART00000158719 |
six4a
|
SIX homeobox 4a |
| chr2_-_195129 | 0.18 |
ENSDART00000126704
|
zgc:113518
|
zgc:113518 |
| chr16_+_35917621 | 0.18 |
|
|
|
| chr9_+_34339522 | 0.16 |
ENSDART00000078051
|
gpr161
|
G protein-coupled receptor 161 |
| chr18_+_16997001 | 0.16 |
ENSDART00000147377
|
si:ch211-218c6.8
|
si:ch211-218c6.8 |
| chr20_-_14116800 | 0.16 |
ENSDART00000152429
|
si:ch211-22i13.2
|
si:ch211-22i13.2 |
| chr16_-_7310925 | 0.14 |
ENSDART00000149030
|
nt5c3a
|
5'-nucleotidase, cytosolic IIIA |
| chr12_+_20578266 | 0.13 |
ENSDART00000016099
|
ENSDARG00000043818
|
ENSDARG00000043818 |
| chr20_-_34767302 | 0.12 |
ENSDART00000033325
|
slc25a24
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 |
| chr24_-_14567403 | 0.08 |
ENSDART00000131830
|
jph1a
|
junctophilin 1a |
| chr2_-_52792076 | 0.08 |
ENSDART00000097716
|
zgc:136336
|
zgc:136336 |
| chr20_-_48677794 | 0.07 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr17_+_37363572 | 0.06 |
ENSDART00000157122
|
elmsan1b
|
ELM2 and Myb/SANT-like domain containing 1b |
| chr19_+_2659309 | 0.05 |
ENSDART00000162293
|
si:ch73-345f18.3
|
si:ch73-345f18.3 |
| chr1_+_54233842 | 0.05 |
ENSDART00000089603
|
golga7ba
|
golgin A7 family, member Ba |
| chr25_-_21796677 | 0.04 |
ENSDART00000089642
|
fbxo31
|
F-box protein 31 |
| KN150703v1_-_47289 | 0.03 |
ENSDART00000168391
|
CABZ01081752.1
|
ENSDARG00000099245 |
| chr16_+_51597530 | 0.03 |
|
|
|
| chr18_-_895821 | 0.02 |
ENSDART00000172518
|
cox5ab
|
cytochrome c oxidase subunit Vab |
| chr5_+_23555868 | 0.01 |
ENSDART00000124379
|
gabarapa
|
GABA(A) receptor-associated protein a |
| chr15_+_37688139 | 0.01 |
ENSDART00000076066
|
lin37
|
lin-37 DREAM MuvB core complex component |
| chr16_+_51597409 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0048855 | post-embryonic morphogenesis(GO:0009886) post-embryonic foregut morphogenesis(GO:0048618) hypophysis morphogenesis(GO:0048850) diencephalon morphogenesis(GO:0048852) adenohypophysis morphogenesis(GO:0048855) |
| 0.2 | 0.5 | GO:0072526 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.1 | 1.0 | GO:0061056 | arterial endothelial cell differentiation(GO:0060842) sclerotome development(GO:0061056) |
| 0.1 | 0.4 | GO:0097704 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) response to oscillatory fluid shear stress(GO:0097702) cellular response to oscillatory fluid shear stress(GO:0097704) |
| 0.1 | 0.7 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.3 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 0.3 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 0.4 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 1.4 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.0 | 0.3 | GO:1901910 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.6 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 0.5 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.2 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.3 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.7 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 1.1 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.4 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 1.1 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.4 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.5 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 1.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.7 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.3 | 1.4 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.2 | 1.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.2 | 0.5 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.5 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 1.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.4 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 1.0 | GO:0005112 | Notch binding(GO:0005112) PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.3 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.3 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 0.7 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.7 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |