Project
DANIO-CODE
Navigation
Downloads
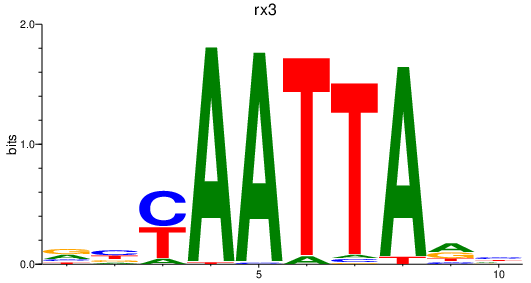
Results for rx3
Z-value: 1.28
Transcription factors associated with rx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rx3
|
ENSDARG00000052893 | retinal homeobox gene 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rx3 | dr10_dc_chr21_+_10663517_10663526 | -0.72 | 1.5e-03 | Click! |
Activity profile of rx3 motif
Sorted Z-values of rx3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of rx3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_40718244 | 6.11 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr11_-_44539778 | 5.42 |
ENSDART00000161846
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr8_+_45326435 | 4.87 |
ENSDART00000134161
|
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr10_-_34058331 | 4.82 |
ENSDART00000046599
|
zar1l
|
zygote arrest 1-like |
| chr13_-_6875290 | 4.71 |
|
|
|
| chr24_+_12689711 | 4.56 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr24_-_14446593 | 4.19 |
|
|
|
| chr17_+_16038358 | 4.02 |
ENSDART00000155336
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr10_-_21404605 | 3.86 |
ENSDART00000125167
|
avd
|
avidin |
| chr24_+_19270877 | 3.24 |
|
|
|
| chr1_-_18118467 | 2.97 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr18_-_43890836 | 2.90 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr2_+_6341404 | 2.72 |
ENSDART00000076700
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr21_-_32027717 | 2.68 |
ENSDART00000131651
|
ENSDARG00000073961
|
ENSDARG00000073961 |
| chr20_-_23527234 | 2.57 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr24_+_12689887 | 2.51 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr10_-_32550351 | 2.46 |
ENSDART00000129395
|
uvrag
|
UV radiation resistance associated gene |
| chr19_+_39689450 | 2.44 |
|
|
|
| chr17_+_16038103 | 2.43 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr4_+_4825461 | 2.31 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr24_-_24999240 | 2.28 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr14_-_8634381 | 2.11 |
ENSDART00000129030
|
zgc:153681
|
zgc:153681 |
| chr12_-_33256671 | 2.07 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr14_+_23420053 | 1.94 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr10_+_6925373 | 1.93 |
ENSDART00000128866
|
ddx4
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 |
| chr6_-_19556029 | 1.90 |
ENSDART00000136019
|
ppp1r12c
|
protein phosphatase 1, regulatory subunit 12C |
| chr12_-_33256754 | 1.86 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr9_-_35824470 | 1.73 |
ENSDART00000140356
|
zp2l1
|
zona pellucida glycoprotein 2, like 1 |
| chr16_+_42567707 | 1.70 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr1_+_18118735 | 1.69 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr25_-_13394261 | 1.68 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr20_-_29961498 | 1.63 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr22_-_21873054 | 1.62 |
|
|
|
| chr1_+_35253862 | 1.60 |
ENSDART00000139636
|
zgc:152968
|
zgc:152968 |
| chr12_-_33257026 | 1.59 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr12_+_33257120 | 1.57 |
|
|
|
| chr20_-_29961589 | 1.54 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr6_+_40925259 | 1.52 |
ENSDART00000002728
ENSDART00000145153 |
eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr18_+_20571460 | 1.47 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr12_-_33256599 | 1.44 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr18_+_8362265 | 1.43 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr10_-_2944190 | 1.40 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
| chr8_-_44247277 | 1.40 |
|
|
|
| chr2_-_15656155 | 1.37 |
ENSDART00000015655
|
tecrl2b
|
trans-2,3-enoyl-CoA reductase-like 2b |
| chr13_-_24778226 | 1.32 |
|
|
|
| chr18_+_20571400 | 1.25 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr11_-_44539726 | 1.21 |
ENSDART00000173360
|
map1lc3c
|
microtubule-associated protein 1 light chain 3 gamma |
| chr14_+_23419864 | 1.17 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr6_-_19556121 | 1.16 |
ENSDART00000136019
|
ppp1r12c
|
protein phosphatase 1, regulatory subunit 12C |
| chr13_-_24778352 | 1.14 |
|
|
|
| chr6_-_19556328 | 1.12 |
ENSDART00000074256
|
ppp1r12c
|
protein phosphatase 1, regulatory subunit 12C |
| chr12_+_22459218 | 1.08 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr13_-_24778261 | 1.08 |
|
|
|
| KN150040v1_-_6426 | 1.01 |
|
|
|
| chr15_-_37037547 | 1.01 |
|
|
|
| chr22_-_551324 | 1.01 |
|
|
|
| chr6_+_59580554 | 1.01 |
|
|
|
| chr20_-_37910887 | 1.00 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr5_+_6391432 | 0.95 |
ENSDART00000170564
ENSDART00000086666 |
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr8_+_45326523 | 0.95 |
ENSDART00000145011
|
pabpc1l
|
poly(A) binding protein, cytoplasmic 1-like |
| chr12_+_22459177 | 0.93 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr20_+_35176069 | 0.90 |
|
|
|
| chr2_+_6341345 | 0.89 |
ENSDART00000058256
|
zp3b
|
zona pellucida glycoprotein 3b |
| chr24_+_16402587 | 0.83 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr15_-_43402935 | 0.77 |
ENSDART00000041677
|
serpine2
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 |
| KN150040v1_+_6648 | 0.73 |
|
|
|
| chr24_-_24999348 | 0.73 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr24_-_24999290 | 0.73 |
ENSDART00000152104
|
phldb2b
|
pleckstrin homology-like domain, family B, member 2b |
| chr22_-_21872864 | 0.72 |
ENSDART00000158501
|
gna11a
|
guanine nucleotide binding protein (G protein), alpha 11a (Gq class) |
| chr16_+_42567668 | 0.71 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr24_-_14447655 | 0.69 |
|
|
|
| chr8_-_25015215 | 0.68 |
ENSDART00000170511
|
nfyal
|
nuclear transcription factor Y, alpha, like |
| chr18_+_20571785 | 0.67 |
ENSDART00000040074
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr14_-_6901783 | 0.66 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr16_-_42105636 | 0.66 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr24_+_12689972 | 0.66 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr4_+_4825628 | 0.65 |
ENSDART00000150309
|
ptprz1b
|
protein tyrosine phosphatase, receptor-type, Z polypeptide 1b |
| chr24_-_14446522 | 0.62 |
|
|
|
| chr14_+_8634323 | 0.61 |
ENSDART00000159920
|
rps6kal
|
ribosomal protein S6 kinase a, like |
| chr17_+_8642373 | 0.59 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr9_-_50304120 | 0.58 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr24_-_14447519 | 0.56 |
|
|
|
| chr3_+_32234155 | 0.56 |
ENSDART00000155967
|
ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr9_-_50304152 | 0.50 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr10_+_6925975 | 0.50 |
|
|
|
| chr5_-_54492042 | 0.49 |
ENSDART00000158007
|
CU467961.1
|
ENSDARG00000078270 |
| chr12_+_22459371 | 0.48 |
ENSDART00000171725
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr11_-_29520809 | 0.47 |
ENSDART00000079117
|
plekha3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr17_+_8642428 | 0.45 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr18_+_8362131 | 0.44 |
ENSDART00000092053
|
chkb
|
choline kinase beta |
| chr17_-_41000670 | 0.43 |
ENSDART00000124715
|
si:dkey-16j16.4
|
si:dkey-16j16.4 |
| chr11_+_34259003 | 0.41 |
|
|
|
| chr16_-_42105733 | 0.40 |
ENSDART00000102798
|
zp3d.2
|
zona pellucida glycoprotein 3d tandem duplicate 2 |
| chr6_+_59580339 | 0.38 |
|
|
|
| chr17_+_8642686 | 0.32 |
ENSDART00000105326
|
tonsl
|
tonsoku-like, DNA repair protein |
| chr14_+_23419894 | 0.30 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr2_+_50873843 | 0.28 |
|
|
|
| chr24_+_12690117 | 0.27 |
ENSDART00000114762
|
nanog
|
nanog homeobox |
| chr7_-_68229115 | 0.26 |
|
|
|
| chr5_-_66441778 | 0.25 |
ENSDART00000168245
|
rps6ka4
|
ribosomal protein S6 kinase, polypeptide 4 |
| chr13_-_25689369 | 0.24 |
ENSDART00000077612
|
rel
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr20_-_29961621 | 0.17 |
ENSDART00000132278
|
rnf144ab
|
ring finger protein 144ab |
| chr22_+_20141311 | 0.17 |
|
|
|
| chr1_+_35253821 | 0.13 |
ENSDART00000179634
|
zgc:152968
|
zgc:152968 |
| chr22_-_20141216 | 0.13 |
ENSDART00000128023
|
btbd2a
|
BTB (POZ) domain containing 2a |
| chr2_-_22162431 | 0.11 |
ENSDART00000135230
|
chd7
|
chromodomain helicase DNA binding protein 7 |
| chr10_-_2944221 | 0.08 |
ENSDART00000132526
|
marveld2a
|
MARVEL domain containing 2a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 8.0 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 0.7 | 3.4 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.4 | 6.6 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.3 | 4.7 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.2 | 1.9 | GO:0061458 | gonad development(GO:0008406) reproductive structure development(GO:0048608) reproductive system development(GO:0061458) |
| 0.2 | 1.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 1.5 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 3.0 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.1 | 7.0 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.1 | 2.4 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.5 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.1 | 3.3 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) positive regulation of proteolysis involved in cellular protein catabolic process(GO:1903052) |
| 0.1 | 3.4 | GO:0031398 | positive regulation of protein ubiquitination(GO:0031398) |
| 0.1 | 1.4 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 0.6 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) DNA damage response, signal transduction resulting in transcription(GO:0042772) |
| 0.0 | 1.4 | GO:0042761 | very long-chain fatty acid biosynthetic process(GO:0042761) |
| 0.0 | 0.3 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 1.5 | GO:0007605 | sensory perception of sound(GO:0007605) |
| 0.0 | 3.7 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 0.7 | GO:0001508 | action potential(GO:0001508) |
| 0.0 | 2.9 | GO:0017148 | negative regulation of translation(GO:0017148) |
| 0.0 | 1.2 | GO:0072329 | monocarboxylic acid catabolic process(GO:0072329) |
| 0.0 | 0.1 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 4.8 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 0.2 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.3 | 3.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.2 | 6.6 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 7.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 1.4 | GO:0035101 | FACT complex(GO:0035101) |
| 0.1 | 1.9 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.7 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 2.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 3.4 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 3.3 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.6 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 1.0 | 7.0 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.8 | 3.9 | GO:0009374 | biotin binding(GO:0009374) |
| 0.6 | 1.7 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.5 | 5.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.4 | 3.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.4 | 3.6 | GO:0035804 | structural constituent of egg coat(GO:0035804) |
| 0.1 | 1.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.1 | 0.5 | GO:0035620 | ceramide transporter activity(GO:0035620) sphingolipid transporter activity(GO:0046624) ceramide 1-phosphate binding(GO:1902387) ceramide 1-phosphate transporter activity(GO:1902388) |
| 0.1 | 3.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 6.6 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.1 | 0.7 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.1 | 3.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 3.3 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.6 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 8.0 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 4.2 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 1.4 | GO:0016627 | oxidoreductase activity, acting on the CH-CH group of donors(GO:0016627) |
| 0.0 | 3.0 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.7 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 1.2 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 1.5 | GO:0000149 | SNARE binding(GO:0000149) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.4 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 7.0 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.2 | 1.9 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.4 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.1 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |