Project
DANIO-CODE
Navigation
Downloads
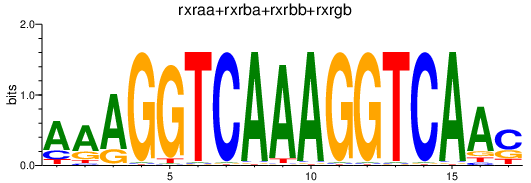
Results for rxraa+rxrba+rxrbb+rxrgb
Z-value: 0.50
Transcription factors associated with rxraa+rxrba+rxrbb+rxrgb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rxrbb
|
ENSDARG00000002006 | retinoid x receptor, beta b |
|
rxrgb
|
ENSDARG00000004697 | retinoid X receptor, gamma b |
|
rxraa
|
ENSDARG00000057737 | retinoid X receptor, alpha a |
|
rxrba
|
ENSDARG00000078954 | retinoid x receptor, beta a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| rxrgb | dr10_dc_chr20_+_33972327_33972354 | 0.66 | 5.6e-03 | Click! |
| rxraa | dr10_dc_chr21_+_17731439_17731676 | 0.62 | 1.0e-02 | Click! |
| rxrbb | dr10_dc_chr16_+_18729942_18730054 | 0.34 | 2.0e-01 | Click! |
| rxrba | dr10_dc_chr19_-_7354071_7354176 | 0.03 | 9.2e-01 | Click! |
Activity profile of rxraa+rxrba+rxrbb+rxrgb motif
Sorted Z-values of rxraa+rxrba+rxrbb+rxrgb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of rxraa+rxrba+rxrbb+rxrgb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_933677 | 1.49 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr1_+_25157675 | 1.47 |
ENSDART00000136984
|
fabp2
|
fatty acid binding protein 2, intestinal |
| chr3_+_39425125 | 1.29 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr13_-_39033893 | 1.18 |
ENSDART00000045434
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr5_-_1325831 | 0.91 |
|
|
|
| chr6_-_13654186 | 0.76 |
ENSDART00000150102
ENSDART00000041269 |
cryba2a
|
crystallin, beta A2a |
| chr16_+_29716279 | 0.74 |
ENSDART00000137153
|
tmod4
|
tropomodulin 4 (muscle) |
| chr25_+_21732255 | 0.69 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr16_+_24697776 | 0.68 |
|
|
|
| chr11_+_11217547 | 0.66 |
ENSDART00000087105
|
myom2a
|
myomesin 2a |
| chr15_-_35563367 | 0.66 |
|
|
|
| chr11_-_42096921 | 0.63 |
|
|
|
| chr5_-_21520111 | 0.62 |
|
|
|
| chr17_-_31642673 | 0.60 |
ENSDART00000030448
|
vsx2
|
visual system homeobox 2 |
| chr19_-_19290260 | 0.57 |
|
|
|
| chr17_+_443558 | 0.54 |
ENSDART00000171386
|
zgc:194887
|
zgc:194887 |
| chr15_+_9321225 | 0.54 |
ENSDART00000055554
|
slc37a4a
|
solute carrier family 37 (glucose-6-phosphate transporter), member 4a |
| chr19_-_22983970 | 0.52 |
ENSDART00000160153
|
pleca
|
plectin a |
| chr15_+_3296905 | 0.52 |
ENSDART00000171723
|
foxo1a
|
forkhead box O1 a |
| chr21_-_26459113 | 0.51 |
ENSDART00000157255
|
cd248b
|
CD248 molecule, endosialin b |
| chr16_-_24697750 | 0.50 |
ENSDART00000163305
|
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr11_-_23079615 | 0.49 |
ENSDART00000125024
|
golt1a
|
golgi transport 1A |
| chr22_+_26543146 | 0.48 |
|
|
|
| chr22_-_23641813 | 0.48 |
ENSDART00000159622
|
cfh
|
complement factor H |
| chr13_-_39033849 | 0.47 |
ENSDART00000045434
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr20_+_23643050 | 0.46 |
ENSDART00000168690
|
palld
|
palladin, cytoskeletal associated protein |
| chr9_+_33167554 | 0.46 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr6_+_60060297 | 0.43 |
ENSDART00000178621
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr18_-_48498261 | 0.42 |
ENSDART00000146346
|
kcnj1a.6
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 6 |
| chr12_-_31342432 | 0.42 |
ENSDART00000148603
|
acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr1_+_23093114 | 0.42 |
|
|
|
| chr8_+_29751639 | 0.41 |
|
|
|
| chr24_-_21027589 | 0.40 |
ENSDART00000154259
|
atp6v1ab
|
ATPase, H+ transporting, lysosomal, V1 subunit Ab |
| chr7_+_36267647 | 0.39 |
ENSDART00000173653
|
chd9
|
chromodomain helicase DNA binding protein 9 |
| chr16_-_46678676 | 0.38 |
ENSDART00000131485
ENSDART00000122584 ENSDART00000132926 |
tmem176l.2
|
transmembrane protein 176l.2 |
| chr4_-_16461748 | 0.38 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| chr21_-_45852590 | 0.36 |
ENSDART00000169816
|
galnt10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr5_-_47519273 | 0.36 |
|
|
|
| chr12_-_30897542 | 0.34 |
ENSDART00000145967
|
tcf7l2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr19_+_177005 | 0.33 |
ENSDART00000111580
|
tmem65
|
transmembrane protein 65 |
| chr3_-_18642744 | 0.33 |
ENSDART00000134208
|
hagh
|
hydroxyacylglutathione hydrolase |
| chr2_-_49060868 | 0.32 |
|
|
|
| chr16_-_23882488 | 0.31 |
ENSDART00000077834
|
rps27.2
|
ribosomal protein S27, isoform 2 |
| chr9_-_41982635 | 0.30 |
ENSDART00000144573
|
obsl1b
|
obscurin-like 1b |
| chr22_+_34454253 | 0.30 |
ENSDART00000156615
|
amigo3
|
adhesion molecule with Ig-like domain 3 |
| chr19_-_22984081 | 0.30 |
ENSDART00000160153
|
pleca
|
plectin a |
| chr5_+_24569070 | 0.30 |
ENSDART00000012268
|
mrpl41
|
mitochondrial ribosomal protein L41 |
| chr12_-_30897752 | 0.28 |
ENSDART00000145967
|
tcf7l2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr13_-_37001997 | 0.28 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr22_-_17570275 | 0.28 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr9_-_48152388 | 0.28 |
|
|
|
| chr1_+_11485480 | 0.27 |
ENSDART00000132560
|
stra6l
|
STRA6-like |
| chr5_-_3562233 | 0.27 |
ENSDART00000143250
|
si:ch1073-189o9.1
|
si:ch1073-189o9.1 |
| chr2_-_32572545 | 0.27 |
ENSDART00000056641
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr5_-_63031745 | 0.26 |
|
|
|
| chr17_-_16079935 | 0.25 |
|
|
|
| chr6_+_40953885 | 0.24 |
ENSDART00000156660
|
patz1
|
POZ (BTB) and AT hook containing zinc finger 1 |
| chr13_-_22713071 | 0.24 |
ENSDART00000142738
|
pbld
|
phenazine biosynthesis like protein domain containing |
| chr22_+_26991665 | 0.24 |
ENSDART00000112826
|
tmem186
|
transmembrane protein 186 |
| chr24_+_13780553 | 0.23 |
ENSDART00000138119
ENSDART00000134221 |
eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr16_-_1479139 | 0.22 |
ENSDART00000036348
|
sim1a
|
single-minded family bHLH transcription factor 1a |
| chr15_+_6870469 | 0.19 |
|
|
|
| chr13_-_41420815 | 0.19 |
ENSDART00000163331
ENSDART00000164732 |
pcdh15a
|
protocadherin-related 15a |
| chr5_-_27394386 | 0.19 |
ENSDART00000171611
|
ppp3cca
|
protein phosphatase 3, catalytic subunit, gamma isozyme, a |
| chr17_-_45750643 | 0.19 |
ENSDART00000074873
|
arf6b
|
ADP-ribosylation factor 6b |
| chr9_+_33167471 | 0.18 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr17_-_45750693 | 0.18 |
ENSDART00000074873
|
arf6b
|
ADP-ribosylation factor 6b |
| chr23_-_45074750 | 0.17 |
ENSDART00000148669
|
si:ch73-269m23.5
|
si:ch73-269m23.5 |
| chr25_-_34608926 | 0.17 |
ENSDART00000130395
|
hist1h4l
|
histone 1, H4, like |
| chr6_+_28127514 | 0.17 |
|
|
|
| chr18_+_38793474 | 0.16 |
ENSDART00000059208
|
fam214a
|
family with sequence similarity 214, member A |
| chr8_+_7974156 | 0.16 |
|
|
|
| chr6_+_28127444 | 0.15 |
|
|
|
| chr23_+_28450814 | 0.15 |
ENSDART00000014983
|
zgc:153867
|
zgc:153867 |
| chr13_+_21695882 | 0.15 |
ENSDART00000165150
|
zswim8
|
zinc finger, SWIM-type containing 8 |
| KN150477v1_+_13133 | 0.14 |
|
|
|
| chr23_+_37535830 | 0.14 |
|
|
|
| chr2_-_56896934 | 0.14 |
ENSDART00000164086
|
slc25a42
|
solute carrier family 25, member 42 |
| chr5_-_55745987 | 0.13 |
ENSDART00000083079
|
acaca
|
acetyl-CoA carboxylase alpha |
| chr9_+_28301359 | 0.12 |
|
|
|
| chr4_+_2725218 | 0.12 |
ENSDART00000007638
|
bcap29
|
B-cell receptor-associated protein 29 |
| chr23_+_8862155 | 0.11 |
ENSDART00000132992
|
sox18
|
SRY (sex determining region Y)-box 18 |
| chr17_-_9774006 | 0.11 |
ENSDART00000149640
|
egln3
|
egl-9 family hypoxia-inducible factor 3 |
| chr7_+_7261692 | 0.11 |
ENSDART00000102629
|
nit1
|
nitrilase 1 |
| chr13_+_45843567 | 0.10 |
ENSDART00000005195
ENSDART00000074547 |
bsdc1
|
BSD domain containing 1 |
| chr7_-_23775835 | 0.09 |
ENSDART00000144616
|
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr13_+_45843396 | 0.09 |
ENSDART00000005195
ENSDART00000074547 |
bsdc1
|
BSD domain containing 1 |
| chr9_-_30752583 | 0.09 |
|
|
|
| chr24_-_7558008 | 0.08 |
ENSDART00000093163
|
galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr2_+_23600656 | 0.08 |
|
|
|
| chr1_+_49894685 | 0.07 |
ENSDART00000020412
|
pkd2
|
polycystic kidney disease 2 |
| chr10_-_30006277 | 0.07 |
ENSDART00000099983
|
bsx
|
brain-specific homeobox |
| chr9_-_29769100 | 0.07 |
ENSDART00000140876
|
cenpj
|
centromere protein J |
| chr7_-_41116672 | 0.06 |
ENSDART00000051679
|
prkdc
|
protein kinase, DNA-activated, catalytic polypeptide |
| chr6_+_12972934 | 0.05 |
ENSDART00000036927
|
ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr12_+_26579396 | 0.04 |
ENSDART00000144355
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr16_+_28667291 | 0.04 |
ENSDART00000018235
|
crot
|
carnitine O-octanoyltransferase |
| chr19_-_22984023 | 0.03 |
ENSDART00000160153
|
pleca
|
plectin a |
| chr17_+_28664644 | 0.02 |
|
|
|
| chr25_-_34635697 | 0.02 |
ENSDART00000113870
|
ENSDARG00000075379
|
ENSDARG00000075379 |
| chr15_+_36355348 | 0.01 |
|
|
|
| chr25_+_20021806 | 0.01 |
ENSDART00000104304
|
bpgm
|
2,3-bisphosphoglycerate mutase |
| chr21_+_302452 | 0.00 |
ENSDART00000151613
|
lhfpl2a
|
lipoma HMGIC fusion partner-like 2a |
| chr6_+_12972739 | 0.00 |
ENSDART00000036927
|
ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.2 | 0.6 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.1 | 0.4 | GO:1903959 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) regulation of organic acid transport(GO:0032890) positive regulation of organic acid transport(GO:0032892) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transport(GO:1903793) regulation of anion transmembrane transport(GO:1903959) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.1 | 0.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.4 | GO:0071549 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) glycerol biosynthetic process from pyruvate(GO:0046327) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.1 | 0.7 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.1 | 1.3 | GO:0030388 | thigmotaxis(GO:0001966) fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.3 | GO:0009438 | methylglyoxal metabolic process(GO:0009438) |
| 0.1 | 0.2 | GO:0035776 | pronephric proximal tubule development(GO:0035776) |
| 0.1 | 0.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.0 | 0.6 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.3 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.0 | 0.5 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 3.5 | GO:0033993 | response to lipid(GO:0033993) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.3 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.0 | 0.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.1 | GO:0044246 | regulation of collagen metabolic process(GO:0010712) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) response to oscillatory fluid shear stress(GO:0097702) cellular response to oscillatory fluid shear stress(GO:0097704) |
| 0.0 | 0.3 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.0 | 0.4 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.0 | 0.1 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.4 | GO:0010107 | potassium ion import(GO:0010107) potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.8 | GO:0007601 | visual perception(GO:0007601) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.7 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.4 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 1.7 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.2 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.7 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.2 | 0.5 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 1.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.4 | GO:0004613 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.1 | 0.3 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.1 | 0.7 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.1 | 0.9 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.0 | 0.2 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.1 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.0 | GO:0016415 | octanoyltransferase activity(GO:0016415) |
| 0.0 | 0.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.6 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |