Project
DANIO-CODE
Navigation
Downloads
Results for rxrab
Z-value: 1.21
Transcription factors associated with rxrab
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
rxrab
|
ENSDARG00000035127 | retinoid x receptor, alpha b |
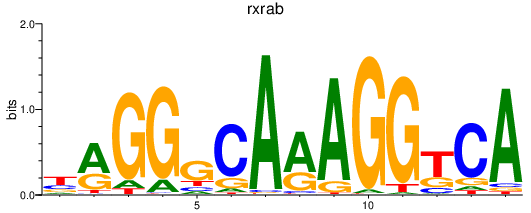
Activity profile of rxrab motif
Sorted Z-values of rxrab motif
Network of associatons between targets according to the STRING database.
First level regulatory network of rxrab
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_933677 | 6.07 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr1_+_25157675 | 5.05 |
ENSDART00000136984
|
fabp2
|
fatty acid binding protein 2, intestinal |
| chr22_-_17570306 | 4.57 |
ENSDART00000139361
|
gpx4a
|
glutathione peroxidase 4a |
| chr12_-_5085227 | 4.33 |
ENSDART00000160729
|
rbp4
|
retinol binding protein 4, plasma |
| chr8_+_31426328 | 3.80 |
ENSDART00000135101
|
sepp1a
|
selenoprotein P, plasma, 1a |
| chr17_+_443558 | 3.73 |
ENSDART00000171386
|
zgc:194887
|
zgc:194887 |
| chr3_-_31672763 | 3.37 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr25_+_5162045 | 3.12 |
ENSDART00000169540
|
ENSDARG00000101164
|
ENSDARG00000101164 |
| chr18_-_17426513 | 2.93 |
ENSDART00000150077
|
ces2
|
carboxylesterase 2 (intestine, liver) |
| KN149726v1_+_1094 | 2.81 |
|
|
|
| chr2_-_43110857 | 2.74 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr5_-_36237656 | 2.70 |
ENSDART00000032481
|
ckma
|
creatine kinase, muscle a |
| chr23_+_44817648 | 2.67 |
ENSDART00000143688
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr25_+_21732255 | 2.65 |
ENSDART00000027393
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr13_+_28689749 | 2.63 |
ENSDART00000101653
|
CU639469.1
|
ENSDARG00000062790 |
| chr10_-_44180588 | 2.63 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr5_+_8415025 | 2.59 |
ENSDART00000046440
|
agpat9l
|
1-acylglycerol-3-phosphate O-acyltransferase 9, like |
| chr5_+_40722565 | 2.59 |
ENSDART00000097546
|
arid3c
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr11_-_35501227 | 2.53 |
ENSDART00000026017
|
bhlhe40
|
basic helix-loop-helix family, member e40 |
| chr13_-_37001997 | 2.51 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr16_-_24697750 | 2.50 |
ENSDART00000163305
|
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr3_+_39425125 | 2.47 |
ENSDART00000146867
|
aldoaa
|
aldolase a, fructose-bisphosphate, a |
| chr16_-_39181608 | 2.43 |
ENSDART00000075517
|
gdf6a
|
growth differentiation factor 6a |
| chr6_+_59788008 | 2.43 |
|
|
|
| chr7_+_38626646 | 2.39 |
ENSDART00000172251
ENSDART00000164019 |
creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr16_+_42925950 | 2.36 |
ENSDART00000159730
ENSDART00000014956 |
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr12_+_27370834 | 2.31 |
ENSDART00000105661
|
meox1
|
mesenchyme homeobox 1 |
| chr5_-_1325831 | 2.29 |
|
|
|
| chr14_-_49115338 | 2.29 |
ENSDART00000056712
|
etfdh
|
electron-transferring-flavoprotein dehydrogenase |
| chr25_+_35797357 | 2.27 |
ENSDART00000152449
|
CR354435.1
|
ENSDARG00000039547 |
| chr4_-_16461748 | 2.24 |
ENSDART00000128835
|
wu:fc23c09
|
wu:fc23c09 |
| chr19_-_19290260 | 2.19 |
|
|
|
| chr17_+_12254292 | 2.16 |
ENSDART00000160722
|
khk
|
ketohexokinase |
| chr21_+_20734431 | 2.14 |
ENSDART00000079732
|
oxct1b
|
3-oxoacid CoA transferase 1b |
| chr23_+_27985224 | 2.13 |
ENSDART00000171859
|
ENSDARG00000100606
|
ENSDARG00000100606 |
| chr16_+_24697776 | 2.13 |
|
|
|
| chr19_+_41274734 | 2.09 |
ENSDART00000126470
|
zgc:85777
|
zgc:85777 |
| chr21_-_18969970 | 1.96 |
ENSDART00000080269
|
pgam2
|
phosphoglycerate mutase 2 (muscle) |
| chr16_-_29779166 | 1.94 |
ENSDART00000067854
|
tnfaip8l2b
|
tumor necrosis factor, alpha-induced protein 8-like 2b |
| chr6_-_39315024 | 1.94 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr6_-_30876091 | 1.93 |
ENSDART00000155330
|
pde4ba
|
phosphodiesterase 4B, cAMP-specific a |
| chr14_-_48951428 | 1.93 |
ENSDART00000157785
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr11_+_24720057 | 1.90 |
ENSDART00000145647
|
sulf2a
|
sulfatase 2a |
| chr12_+_27032862 | 1.90 |
|
|
|
| chr9_+_33167554 | 1.87 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr15_+_19902697 | 1.84 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr16_-_32022177 | 1.84 |
ENSDART00000139664
|
styk1
|
serine/threonine/tyrosine kinase 1 |
| chr14_+_35906366 | 1.84 |
ENSDART00000105602
|
elovl6
|
ELOVL fatty acid elongase 6 |
| chr4_-_2615160 | 1.84 |
ENSDART00000140760
|
e2f7
|
E2F transcription factor 7 |
| chr20_+_30894667 | 1.82 |
ENSDART00000145066
|
nhsl1b
|
NHS-like 1b |
| chr22_+_38276148 | 1.82 |
ENSDART00000104504
|
si:ch211-284e20.8
|
si:ch211-284e20.8 |
| chr5_+_37053530 | 1.76 |
ENSDART00000161051
|
sptbn2
|
spectrin, beta, non-erythrocytic 2 |
| chr12_+_31558667 | 1.74 |
ENSDART00000152971
|
dnmbp
|
dynamin binding protein |
| chr5_-_64315770 | 1.72 |
ENSDART00000169287
|
zgc:110283
|
zgc:110283 |
| chr25_+_4490129 | 1.71 |
|
|
|
| chr2_+_53253623 | 1.70 |
ENSDART00000121980
|
creb3l3b
|
cAMP responsive element binding protein 3-like 3b |
| chr11_-_18166056 | 1.64 |
ENSDART00000155752
|
sfmbt1
|
Scm-like with four mbt domains 1 |
| chr19_+_32579358 | 1.63 |
ENSDART00000021798
|
fabp11a
|
fatty acid binding protein 11a |
| chr25_+_15551474 | 1.62 |
ENSDART00000137375
|
spon1b
|
spondin 1b |
| chr20_+_13279404 | 1.62 |
ENSDART00000025644
|
ppp2r5a
|
protein phosphatase 2, regulatory subunit B', alpha isoform |
| chr3_-_56474209 | 1.58 |
ENSDART00000156398
|
si:ch211-189a21.1
|
si:ch211-189a21.1 |
| chr9_-_1969197 | 1.57 |
ENSDART00000080608
|
hoxd10a
|
homeobox D10a |
| chr14_+_6122948 | 1.56 |
ENSDART00000149783
ENSDART00000148461 |
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr3_-_24989269 | 1.56 |
ENSDART00000154724
|
chadla
|
chondroadherin-like a |
| chr10_+_5693933 | 1.53 |
ENSDART00000159769
|
pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr11_+_11217547 | 1.51 |
ENSDART00000087105
|
myom2a
|
myomesin 2a |
| chr2_+_48448974 | 1.49 |
ENSDART00000023040
|
hes6
|
hes family bHLH transcription factor 6 |
| chr21_+_25199691 | 1.47 |
ENSDART00000168140
ENSDART00000112783 |
tmem45b
|
transmembrane protein 45B |
| chr7_+_31567166 | 1.45 |
ENSDART00000099785
ENSDART00000122506 |
mybpc3
|
myosin binding protein C, cardiac |
| chr3_-_18642744 | 1.45 |
ENSDART00000134208
|
hagh
|
hydroxyacylglutathione hydrolase |
| chr12_-_31342432 | 1.44 |
ENSDART00000148603
|
acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr2_-_39052185 | 1.44 |
ENSDART00000048838
|
rbp2b
|
retinol binding protein 2b, cellular |
| chr8_-_12171624 | 1.32 |
ENSDART00000132824
|
dab2ipa
|
DAB2 interacting protein a |
| chr23_+_32409339 | 1.31 |
ENSDART00000149698
|
slc39a5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr3_-_45420882 | 1.30 |
ENSDART00000161507
|
zgc:153426
|
zgc:153426 |
| chr5_+_18827478 | 1.30 |
ENSDART00000089078
|
acacb
|
acetyl-CoA carboxylase beta |
| chr25_+_30701751 | 1.29 |
|
|
|
| chr15_-_20297270 | 1.23 |
ENSDART00000123910
|
ppp1r14ab
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ab |
| chr20_+_15119519 | 1.23 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr12_-_34612821 | 1.23 |
ENSDART00000153272
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr9_-_41982635 | 1.22 |
ENSDART00000144573
|
obsl1b
|
obscurin-like 1b |
| chr11_+_30482530 | 1.22 |
ENSDART00000103270
|
slc22a7a
|
solute carrier family 22 (organic anion transporter), member 7a |
| chr17_+_51816782 | 1.22 |
ENSDART00000159072
|
flvcr2a
|
feline leukemia virus subgroup C cellular receptor family, member 2a |
| chr23_+_27141681 | 1.21 |
ENSDART00000054238
|
mipa
|
major intrinsic protein of lens fiber a |
| chr7_-_31170266 | 1.21 |
ENSDART00000075398
|
cilp
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr9_-_22347397 | 1.19 |
ENSDART00000146188
|
crygm2d16
|
crystallin, gamma M2d16 |
| chr23_+_44951868 | 1.19 |
|
|
|
| chr5_-_29891016 | 1.18 |
ENSDART00000138464
|
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr15_+_3296905 | 1.18 |
ENSDART00000171723
|
foxo1a
|
forkhead box O1 a |
| chr19_-_20508993 | 1.17 |
ENSDART00000129917
|
mpp6b
|
membrane protein, palmitoylated 6b (MAGUK p55 subfamily member 6) |
| chr6_+_60060297 | 1.16 |
ENSDART00000178621
|
pck1
|
phosphoenolpyruvate carboxykinase 1 (soluble) |
| chr23_-_624534 | 1.16 |
ENSDART00000132175
|
nadl1.1
|
neural adhesion molecule L1.1 |
| chr7_-_23775835 | 1.15 |
ENSDART00000144616
|
dhrs4
|
dehydrogenase/reductase (SDR family) member 4 |
| chr25_-_34608926 | 1.12 |
ENSDART00000130395
|
hist1h4l
|
histone 1, H4, like |
| chr16_-_22218191 | 1.11 |
ENSDART00000140175
|
si:dkey-71b5.3
|
si:dkey-71b5.3 |
| chr20_-_53162473 | 1.10 |
ENSDART00000164460
|
gata4
|
GATA binding protein 4 |
| chr21_-_12958161 | 1.09 |
ENSDART00000133517
|
MYORG
|
si:dkey-228b2.5 |
| chr12_-_34612758 | 1.09 |
ENSDART00000153272
|
bahcc1b
|
BAH domain and coiled-coil containing 1b |
| chr23_+_42370612 | 1.05 |
ENSDART00000161812
|
cyp2aa9
|
cytochrome P450, family 2, subfamily AA, polypeptide 9 |
| chr10_-_30016761 | 1.04 |
ENSDART00000078800
|
lim2.1
|
lens intrinsic membrane protein 2.1 |
| chr19_+_32570656 | 1.04 |
ENSDART00000005255
|
mrpl53
|
mitochondrial ribosomal protein L53 |
| chr21_-_45852590 | 1.01 |
ENSDART00000169816
|
galnt10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
| chr20_+_15119754 | 1.01 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr22_-_26971466 | 1.01 |
ENSDART00000087202
|
ENSDARG00000061256
|
ENSDARG00000061256 |
| chr5_-_63031745 | 0.99 |
|
|
|
| chr15_-_47264693 | 0.98 |
|
|
|
| chr5_-_21520111 | 0.98 |
|
|
|
| chr10_+_31358236 | 0.98 |
ENSDART00000145562
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr17_-_16079935 | 0.97 |
|
|
|
| chr16_+_55167489 | 0.97 |
ENSDART00000149795
|
nr0b2a
|
nuclear receptor subfamily 0, group B, member 2a |
| chr21_+_13063614 | 0.95 |
|
|
|
| chr22_-_20101177 | 0.95 |
ENSDART00000138688
|
creb3l3a
|
cAMP responsive element binding protein 3-like 3a |
| chr19_+_43525255 | 0.94 |
|
|
|
| chr19_+_11061965 | 0.94 |
ENSDART00000142975
|
si:ch1073-70f20.1
|
si:ch1073-70f20.1 |
| chr1_+_50348973 | 0.93 |
|
|
|
| chr5_+_26804344 | 0.93 |
ENSDART00000121886
ENSDART00000005025 |
hdr
|
hematopoietic death receptor |
| chr21_+_15737517 | 0.93 |
ENSDART00000151161
|
ENSDARG00000044755
|
ENSDARG00000044755 |
| chr19_+_43525673 | 0.92 |
|
|
|
| chr23_-_3814291 | 0.91 |
|
|
|
| chr9_+_1968455 | 0.90 |
|
|
|
| chr16_-_8711133 | 0.89 |
|
|
|
| chr10_-_17484971 | 0.88 |
|
|
|
| chr18_+_17622557 | 0.86 |
ENSDART00000046891
|
cetp
|
cholesteryl ester transfer protein, plasma |
| chr15_-_16948863 | 0.85 |
ENSDART00000062135
|
ENSDARG00000042379
|
ENSDARG00000042379 |
| chr4_+_25637474 | 0.85 |
ENSDART00000066942
ENSDART00000041965 |
acot15
|
acyl-CoA thioesterase 15 |
| chr20_+_25567046 | 0.84 |
ENSDART00000153071
|
pfas
|
phosphoribosylformylglycinamidine synthase |
| chr14_-_27754549 | 0.84 |
ENSDART00000135337
|
zgc:64189
|
zgc:64189 |
| chr19_+_41628053 | 0.84 |
|
|
|
| chr2_+_46179589 | 0.82 |
ENSDART00000125971
|
gpc1b
|
glypican 1b |
| chr20_-_48773402 | 0.82 |
ENSDART00000161769
|
mgst3a
|
microsomal glutathione S-transferase 3a |
| chr14_-_32449229 | 0.82 |
ENSDART00000172996
|
AL590151.1
|
ENSDARG00000053570 |
| chr8_+_25984588 | 0.81 |
|
|
|
| chr13_-_40600924 | 0.81 |
ENSDART00000099847
ENSDART00000057046 |
st3gal7
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 7 |
| chr1_+_11485480 | 0.80 |
ENSDART00000132560
|
stra6l
|
STRA6-like |
| chr14_-_9216303 | 0.79 |
ENSDART00000054689
|
atoh8
|
atonal bHLH transcription factor 8 |
| chr3_+_12633028 | 0.79 |
ENSDART00000168382
|
cyp2k8
|
cytochrome P450, family 2, subfamily K, polypeptide 8 |
| chr19_-_22914312 | 0.78 |
|
|
|
| chr5_+_37600633 | 0.78 |
ENSDART00000100769
|
hsd20b2
|
hydroxysteroid (20-beta) dehydrogenase 2 |
| chr12_-_34621359 | 0.76 |
|
|
|
| chr25_+_3884499 | 0.75 |
ENSDART00000104926
|
ENSDARG00000058108
|
ENSDARG00000058108 |
| chr17_-_24666272 | 0.75 |
ENSDART00000105457
|
morn2
|
MORN repeat containing 2 |
| chr9_+_28301359 | 0.75 |
|
|
|
| chr16_-_1479139 | 0.73 |
ENSDART00000036348
|
sim1a
|
single-minded family bHLH transcription factor 1a |
| chr18_+_12687027 | 0.73 |
ENSDART00000144246
|
tbxas1
|
thromboxane A synthase 1 (platelet) |
| chr20_-_49307929 | 0.73 |
ENSDART00000057700
|
naa20
|
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
| chr4_+_2725218 | 0.73 |
ENSDART00000007638
|
bcap29
|
B-cell receptor-associated protein 29 |
| chr16_+_46239355 | 0.71 |
ENSDART00000168145
|
bola1
|
bolA family member 1 |
| chr1_+_23093114 | 0.70 |
|
|
|
| chr3_+_33629940 | 0.70 |
ENSDART00000169337
|
ier2a
|
immediate early response 2a |
| chr22_-_17570275 | 0.68 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr25_+_21919595 | 0.67 |
ENSDART00000156517
|
ENSDARG00000062199
|
ENSDARG00000062199 |
| chr7_+_20377678 | 0.66 |
ENSDART00000173710
|
si:dkey-19b23.15
|
si:dkey-19b23.15 |
| chr19_+_4142216 | 0.66 |
|
|
|
| chr1_-_49274993 | 0.66 |
ENSDART00000050603
|
hadh
|
hydroxyacyl-CoA dehydrogenase |
| chr15_-_47264619 | 0.66 |
|
|
|
| chr2_-_49060868 | 0.65 |
|
|
|
| chr13_-_44492897 | 0.65 |
|
|
|
| chr6_-_43892838 | 0.65 |
ENSDART00000148646
ENSDART00000148909 |
foxp1b
|
forkhead box P1b |
| chr20_+_13279491 | 0.65 |
ENSDART00000025644
|
ppp2r5a
|
protein phosphatase 2, regulatory subunit B', alpha isoform |
| chr11_-_30971376 | 0.65 |
ENSDART00000170700
|
nacc1b
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing b |
| chr15_-_1880456 | 0.63 |
ENSDART00000102410
|
taf15
|
TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr2_-_30005139 | 0.63 |
ENSDART00000146968
|
cnpy1
|
canopy1 |
| chr8_+_30654835 | 0.63 |
ENSDART00000138750
|
adora2aa
|
adenosine A2a receptor a |
| chr21_-_4086050 | 0.62 |
ENSDART00000099389
|
dnlz
|
DNL-type zinc finger |
| chr7_-_51186389 | 0.61 |
ENSDART00000174328
|
arhgap6
|
Rho GTPase activating protein 6 |
| chr20_+_49273889 | 0.61 |
ENSDART00000166051
ENSDART00000112689 |
crnkl1
|
crooked neck pre-mRNA splicing factor 1 |
| chr22_-_15694002 | 0.61 |
ENSDART00000105692
|
mrpl54
|
mitochondrial ribosomal protein L54 |
| chr3_+_49166063 | 0.60 |
ENSDART00000156347
|
epn3a
|
epsin 3a |
| chr14_-_6939857 | 0.59 |
ENSDART00000108796
|
stox2b
|
storkhead box 2b |
| chr16_+_5470419 | 0.59 |
|
|
|
| chr17_+_43605118 | 0.59 |
ENSDART00000156271
|
cfap99
|
cilia and flagella associated protein 99 |
| chr13_-_25068717 | 0.58 |
ENSDART00000057605
|
adka
|
adenosine kinase a |
| chr12_+_27035744 | 0.57 |
ENSDART00000025966
|
hoxb6b
|
homeobox B6b |
| chr2_-_6351592 | 0.57 |
|
|
|
| chr18_-_48498261 | 0.56 |
ENSDART00000146346
|
kcnj1a.6
|
potassium inwardly-rectifying channel, subfamily J, member 1a, tandem duplicate 6 |
| chr18_+_16755239 | 0.56 |
ENSDART00000133490
|
lyve1b
|
lymphatic vessel endothelial hyaluronic receptor 1b |
| chr2_+_3992842 | 0.55 |
ENSDART00000103596
ENSDART00000161880 |
npc1
|
Niemann-Pick disease, type C1 |
| chr19_-_22914516 | 0.55 |
|
|
|
| chr22_+_18218979 | 0.55 |
|
|
|
| chr1_-_51086211 | 0.54 |
ENSDART00000045894
|
rnaseh2a
|
ribonuclease H2, subunit A |
| chr16_+_28667291 | 0.54 |
ENSDART00000018235
|
crot
|
carnitine O-octanoyltransferase |
| chr10_-_6453139 | 0.53 |
ENSDART00000168549
|
ca9
|
carbonic anhydrase IX |
| chr11_+_30035395 | 0.53 |
ENSDART00000122756
|
si:dkey-163f14.6
|
si:dkey-163f14.6 |
| chr2_+_23600656 | 0.53 |
|
|
|
| chr21_+_15737192 | 0.53 |
ENSDART00000151161
|
ENSDARG00000044755
|
ENSDARG00000044755 |
| chr12_+_29994880 | 0.51 |
ENSDART00000042572
ENSDART00000153025 |
ablim1b
|
actin binding LIM protein 1b |
| chr21_-_34809442 | 0.51 |
ENSDART00000029708
|
zgc:56585
|
zgc:56585 |
| chr23_+_36241656 | 0.50 |
ENSDART00000011201
|
copz1
|
coatomer protein complex, subunit zeta 1 |
| chr5_-_47519273 | 0.50 |
|
|
|
| chr13_-_37002066 | 0.49 |
ENSDART00000135510
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr18_+_18465885 | 0.49 |
ENSDART00000165079
|
siah1
|
siah E3 ubiquitin protein ligase 1 |
| chr15_-_29900 | 0.49 |
|
|
|
| chr13_+_45843396 | 0.48 |
ENSDART00000005195
ENSDART00000074547 |
bsdc1
|
BSD domain containing 1 |
| chr24_-_21768269 | 0.48 |
ENSDART00000081178
|
c1qtnf9
|
C1q and TNF related 9 |
| chr15_+_494813 | 0.48 |
ENSDART00000155682
|
nipsnap2
|
nipsnap homolog 2 |
| chr6_+_28127514 | 0.47 |
|
|
|
| chr2_-_10602948 | 0.47 |
ENSDART00000016369
|
wls
|
wntless Wnt ligand secretion mediator |
| chr8_+_7974156 | 0.47 |
|
|
|
| chr14_-_48951396 | 0.45 |
ENSDART00000157785
|
rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr1_+_43881022 | 0.45 |
ENSDART00000003022
ENSDART00000137980 |
med19b
|
mediator complex subunit 19b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) |
| 0.8 | 2.5 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.8 | 2.4 | GO:0046552 | blood vessel maturation(GO:0001955) retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.7 | 5.4 | GO:0046314 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.6 | 5.1 | GO:0046864 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.6 | 1.8 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.5 | 2.1 | GO:0046950 | cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.5 | 1.4 | GO:0010747 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) plasma membrane long-chain fatty acid transport(GO:0015911) regulation of organic acid transport(GO:0032890) positive regulation of organic acid transport(GO:0032892) fatty acid transmembrane transport(GO:1902001) positive regulation of anion transport(GO:1903793) regulation of anion transmembrane transport(GO:1903959) positive regulation of anion transmembrane transport(GO:1903961) regulation of fatty acid transport(GO:2000191) positive regulation of fatty acid transport(GO:2000193) |
| 0.4 | 1.3 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.4 | 1.2 | GO:0097037 | heme export(GO:0097037) |
| 0.4 | 1.2 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.4 | 2.7 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.3 | 1.3 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.3 | 2.3 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.3 | 2.2 | GO:0035860 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.3 | 1.2 | GO:0019543 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) glycerol biosynthetic process from pyruvate(GO:0046327) cellular response to dexamethasone stimulus(GO:0071549) |
| 0.3 | 0.9 | GO:0034372 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) reverse cholesterol transport(GO:0043691) phospholipid homeostasis(GO:0055091) |
| 0.3 | 1.8 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 0.2 | 1.0 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.2 | 1.9 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.2 | 2.6 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.2 | 2.2 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.2 | 1.3 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.2 | 1.9 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 0.2 | 2.2 | GO:0009746 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.2 | 1.5 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.2 | 1.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.2 | 0.6 | GO:0098751 | bone cell development(GO:0098751) |
| 0.2 | 0.7 | GO:0035776 | pronephric proximal tubule development(GO:0035776) |
| 0.2 | 0.7 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 1.5 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.2 | 1.2 | GO:0006833 | water transport(GO:0006833) |
| 0.2 | 1.8 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 | 0.7 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.7 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.6 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 2.5 | GO:0030388 | thigmotaxis(GO:0001966) fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 1.8 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 5.0 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 0.5 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.1 | 0.8 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.1 | 2.1 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 2.3 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.1 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.6 | GO:0061469 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.4 | GO:0006751 | glutathione catabolic process(GO:0006751) response to estradiol(GO:0032355) |
| 0.1 | 9.7 | GO:0033993 | response to lipid(GO:0033993) |
| 0.1 | 1.6 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.1 | 0.5 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.1 | 2.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 1.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.3 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 0.8 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.8 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.1 | 2.0 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 1.0 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.1 | 0.4 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.1 | 3.8 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 2.9 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 1.0 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.4 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 3.8 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 1.8 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 1.1 | GO:0030835 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) actin filament capping(GO:0051693) |
| 0.0 | 0.5 | GO:0030032 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.2 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.0 | 0.3 | GO:0035479 | angioblast cell migration from lateral mesoderm to midline(GO:0035479) |
| 0.0 | 0.4 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.1 | GO:0043545 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.4 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 1.3 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.4 | GO:0030073 | insulin secretion(GO:0030073) |
| 0.0 | 0.2 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.6 | GO:0010107 | potassium ion import(GO:0010107) potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 1.2 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 0.9 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.0 | 1.5 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.0 | 1.3 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.6 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.8 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.5 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.5 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 2.7 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 1.5 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.9 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 2.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.4 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.7 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.1 | 2.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.1 | 2.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 0.6 | GO:0000974 | Prp19 complex(GO:0000974) U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 3.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 3.4 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 2.2 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 1.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 1.1 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 3.8 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.6 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.0 | GO:1902737 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 16.6 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 1.9 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.5 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 1.6 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 1.1 | GO:0000786 | nucleosome(GO:0000786) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.3 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 1.0 | 11.1 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.9 | 4.3 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.7 | 5.4 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.5 | 2.1 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.4 | 2.6 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.4 | 1.5 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) amidine-lyase activity(GO:0016842) |
| 0.4 | 2.3 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.3 | 2.0 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.3 | 1.2 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.3 | 2.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.3 | 0.8 | GO:0047291 | lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.3 | 5.0 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 2.7 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.2 | 2.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 1.0 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.2 | 1.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 0.6 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.2 | 2.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.2 | 0.6 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 1.8 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.2 | 0.5 | GO:0016415 | octanoyltransferase activity(GO:0016415) |
| 0.2 | 4.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 1.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.2 | 0.7 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.1 | 1.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 1.4 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 0.4 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 1.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.1 | 2.3 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 1.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.6 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 1.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.8 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 0.5 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.7 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 0.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.8 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.3 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.1 | 0.1 | GO:0000104 | succinate dehydrogenase activity(GO:0000104) |
| 0.1 | 1.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 2.3 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.1 | 2.2 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.1 | 1.0 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.9 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.0 | 1.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 2.4 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 6.7 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 3.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 2.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 2.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 2.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.3 | 2.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.3 | 1.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 1.1 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.1 | 3.1 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.1 | 3.4 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 2.5 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.1 | 1.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.4 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.1 | 0.5 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.8 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.2 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 2.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.3 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 2.2 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |