Project
DANIO-CODE
Navigation
Downloads
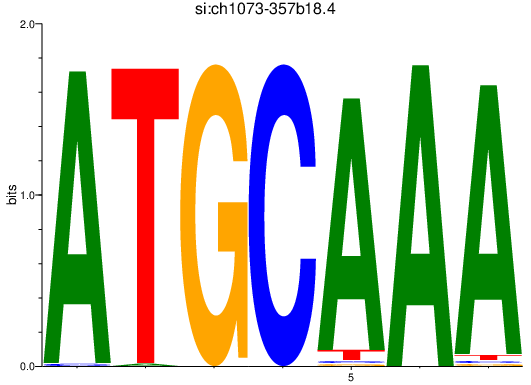
Results for si:ch1073-357b18.4
Z-value: 4.36
Transcription factors associated with si:ch1073-357b18.4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
si_ch1073-357b18.4
|
ENSDARG00000096602 | si_ch1073-357b18.4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:ch1073-357b18.4 | dr10_dc_chr12_-_280962_281088 | 0.68 | 3.8e-03 | Click! |
Activity profile of si:ch1073-357b18.4 motif
Sorted Z-values of si:ch1073-357b18.4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of si:ch1073-357b18.4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_24063849 | 13.57 |
ENSDART00000135084
|
apoa2
|
apolipoprotein A-II |
| chr25_-_32344881 | 13.07 |
ENSDART00000012862
|
isl2a
|
ISL LIM homeobox 2a |
| chr19_-_5441932 | 12.80 |
ENSDART00000105034
|
cyt1l
|
type I cytokeratin, enveloping layer, like |
| chr17_+_19479310 | 11.19 |
ENSDART00000077804
|
slc22a15
|
solute carrier family 22, member 15 |
| chr22_-_37414940 | 10.87 |
ENSDART00000104493
|
sox2
|
SRY (sex determining region Y)-box 2 |
| chr9_-_47041677 | 9.49 |
ENSDART00000054137
|
igfbp5b
|
insulin-like growth factor binding protein 5b |
| chr6_-_39315024 | 9.41 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr2_+_22324846 | 9.31 |
ENSDART00000044371
|
tox
|
thymocyte selection-associated high mobility group box |
| chr25_-_30845998 | 8.82 |
ENSDART00000027661
|
myod1
|
myogenic differentiation 1 |
| chr14_-_24463906 | 8.61 |
ENSDART00000126199
|
slit3
|
slit homolog 3 (Drosophila) |
| chr3_+_23557320 | 8.39 |
ENSDART00000046638
|
hoxb8a
|
homeobox B8a |
| chr23_+_23559246 | 8.38 |
ENSDART00000172214
|
agrn
|
agrin |
| chr4_-_12863235 | 8.20 |
ENSDART00000080536
|
hmga2
|
high mobility group AT-hook 2 |
| chr19_-_5435448 | 7.99 |
ENSDART00000027701
|
krt92
|
keratin 92 |
| chr9_+_54612545 | 7.98 |
ENSDART00000104475
|
tmsb4x
|
thymosin, beta 4 x |
| chr13_+_1595986 | 7.78 |
|
|
|
| chr3_+_23546802 | 7.77 |
ENSDART00000023674
|
hoxb9a
|
homeobox B9a |
| chr9_-_23081918 | 7.69 |
ENSDART00000143888
|
neb
|
nebulin |
| chr25_+_31547276 | 7.31 |
ENSDART00000090727
|
duox
|
dual oxidase |
| chr5_+_37053530 | 7.15 |
ENSDART00000161051
|
sptbn2
|
spectrin, beta, non-erythrocytic 2 |
| chr3_-_6078015 | 7.12 |
ENSDART00000165715
|
BX284638.1
|
ENSDARG00000098850 |
| chr18_+_38768524 | 7.10 |
ENSDART00000143735
|
si:ch211-215d8.2
|
si:ch211-215d8.2 |
| chr23_-_31446156 | 7.00 |
ENSDART00000053367
|
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr4_-_12862869 | 6.99 |
ENSDART00000080536
|
hmga2
|
high mobility group AT-hook 2 |
| chr13_+_28574593 | 6.99 |
ENSDART00000126845
|
ldb1a
|
LIM domain binding 1a |
| chr2_+_55859099 | 6.97 |
ENSDART00000097753
ENSDART00000141688 |
nmrk2
|
nicotinamide riboside kinase 2 |
| chr5_+_12786897 | 6.91 |
|
|
|
| chr16_-_40891298 | 6.88 |
|
|
|
| chr6_+_56163589 | 6.86 |
ENSDART00000150219
|
tfap2c
|
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr13_+_11305781 | 6.83 |
|
|
|
| chr9_-_23081250 | 6.82 |
|
|
|
| chr21_-_28883441 | 6.70 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr14_+_21531709 | 6.63 |
ENSDART00000144367
|
ctbp1
|
C-terminal binding protein 1 |
| chr23_-_9924987 | 6.62 |
ENSDART00000005015
|
prkcbp1l
|
protein kinase C binding protein 1, like |
| chr12_+_7421078 | 6.61 |
ENSDART00000163114
|
phyhiplb
|
phytanoyl-CoA 2-hydroxylase interacting protein-like b |
| chr4_-_8610868 | 6.61 |
ENSDART00000067322
|
fbxl14b
|
F-box and leucine-rich repeat protein 14b |
| chr4_-_12862708 | 6.59 |
ENSDART00000080536
|
hmga2
|
high mobility group AT-hook 2 |
| chr24_-_31772736 | 6.57 |
|
|
|
| chr6_+_28886741 | 6.53 |
ENSDART00000065137
|
tp63
|
tumor protein p63 |
| chr7_+_15623852 | 6.45 |
ENSDART00000161608
|
pax6b
|
paired box 6b |
| chr19_-_22956977 | 6.42 |
ENSDART00000019505
|
pleca
|
plectin a |
| chr7_+_20699191 | 6.38 |
ENSDART00000062003
|
efnb3b
|
ephrin-B3b |
| chr1_+_35933511 | 6.37 |
ENSDART00000010632
|
ednraa
|
endothelin receptor type Aa |
| chr11_+_5745644 | 6.33 |
ENSDART00000179139
|
arid3a
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr10_+_6316944 | 6.32 |
ENSDART00000162428
|
tpm2
|
tropomyosin 2 (beta) |
| chr3_+_34540552 | 6.31 |
ENSDART00000007073
ENSDART00000133457 |
dlx4a
|
distal-less homeobox 4a |
| chr7_-_11812634 | 6.29 |
ENSDART00000101537
|
mex3b
|
mex-3 RNA binding family member B |
| chr15_-_41288480 | 6.28 |
ENSDART00000155359
|
smco4
|
single-pass membrane protein with coiled-coil domains 4 |
| chr21_-_41286846 | 6.28 |
ENSDART00000167339
|
msx2b
|
muscle segment homeobox 2b |
| chr9_-_55204516 | 6.24 |
|
|
|
| chr24_+_24308055 | 6.02 |
|
|
|
| chr1_-_16894589 | 5.99 |
ENSDART00000039917
|
acsl1a
|
acyl-CoA synthetase long-chain family member 1a |
| chr8_+_28584427 | 5.96 |
ENSDART00000097213
|
tcf15
|
transcription factor 15 |
| chr13_+_29640492 | 5.88 |
ENSDART00000160944
|
pax2a
|
paired box 2a |
| chr4_-_20435099 | 5.87 |
ENSDART00000055317
|
lrrc17
|
leucine rich repeat containing 17 |
| chr7_+_19300351 | 5.79 |
ENSDART00000169060
|
si:ch211-212k18.5
|
si:ch211-212k18.5 |
| chr9_+_21911860 | 5.78 |
ENSDART00000102021
|
sox1a
|
SRY (sex determining region Y)-box 1a |
| chr14_-_32624823 | 5.77 |
ENSDART00000114973
|
cdx4
|
caudal type homeobox 4 |
| chr12_-_36415239 | 5.68 |
|
|
|
| chr19_-_5464238 | 5.64 |
ENSDART00000105806
|
cyt1
|
type I cytokeratin, enveloping layer |
| chr16_-_29593569 | 5.63 |
ENSDART00000150028
|
onecutl
|
one cut domain, family member, like |
| chr20_-_53560663 | 5.60 |
ENSDART00000146001
|
wasf1
|
WAS protein family, member 1 |
| chr17_-_44135924 | 5.56 |
ENSDART00000156648
|
otx2
|
orthodenticle homeobox 2 |
| chr14_-_26406720 | 5.56 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr10_+_18994733 | 5.55 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr8_+_17148864 | 5.52 |
ENSDART00000140531
|
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr2_-_18275994 | 5.51 |
ENSDART00000155124
|
ptprfb
|
protein tyrosine phosphatase, receptor type, f, b |
| chr2_+_47864913 | 5.50 |
|
|
|
| chr25_+_19636029 | 5.50 |
ENSDART00000067354
|
zgc:101783
|
zgc:101783 |
| chr24_-_26165778 | 5.48 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr6_-_8501230 | 5.48 |
ENSDART00000143956
|
cavin2b
|
caveolae associated protein 2b |
| chr10_+_22759607 | 5.46 |
|
|
|
| chr15_-_18638643 | 5.41 |
ENSDART00000142010
|
ncam1b
|
neural cell adhesion molecule 1b |
| chr23_-_29138952 | 5.36 |
ENSDART00000002812
|
casz1
|
castor zinc finger 1 |
| chr17_+_33766838 | 5.29 |
ENSDART00000132294
|
fut8a
|
fucosyltransferase 8a (alpha (1,6) fucosyltransferase) |
| chr19_+_3713027 | 5.28 |
ENSDART00000125673
|
nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr10_+_10428297 | 5.25 |
ENSDART00000179214
|
sardh
|
sarcosine dehydrogenase |
| chr15_-_14616083 | 5.25 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr25_-_13580057 | 5.13 |
ENSDART00000090226
|
znf319b
|
zinc finger protein 319b |
| chr14_-_40454194 | 5.08 |
ENSDART00000166621
|
ef1
|
E74-like factor 1 (ets domain transcription factor) |
| chr13_+_19191645 | 5.08 |
ENSDART00000058036
|
emx2
|
empty spiracles homeobox 2 |
| chr1_-_19900799 | 5.07 |
ENSDART00000124770
|
ugt8
|
UDP glycosyltransferase 8 |
| chr2_-_30198789 | 5.03 |
ENSDART00000019149
|
rpl7
|
ribosomal protein L7 |
| chr1_-_6976446 | 5.00 |
ENSDART00000085203
|
efnb2b
|
ephrin-B2b |
| chr24_-_40968409 | 4.97 |
ENSDART00000169315
ENSDART00000171543 |
smyhc1
|
slow myosin heavy chain 1 |
| chr3_-_44113070 | 4.96 |
ENSDART00000160717
|
znf750
|
zinc finger protein 750 |
| chr7_-_52283383 | 4.96 |
ENSDART00000165649
|
tcf12
|
transcription factor 12 |
| chr13_-_1980818 | 4.94 |
ENSDART00000110814
|
fam83b
|
family with sequence similarity 83, member B |
| chr7_-_33558939 | 4.89 |
ENSDART00000074729
|
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr1_+_30992553 | 4.88 |
ENSDART00000112333
|
cnnm2b
|
cyclin and CBS domain divalent metal cation transport mediator 2b |
| chr24_+_21200975 | 4.86 |
ENSDART00000126519
|
shisa2b
|
shisa family member 2b |
| chr3_+_12287067 | 4.86 |
ENSDART00000158060
|
vasnb
|
vasorin b |
| chr23_+_10552781 | 4.85 |
|
|
|
| chr13_+_11305846 | 4.84 |
|
|
|
| chr9_-_54126121 | 4.77 |
ENSDART00000126314
|
pcdh8
|
protocadherin 8 |
| chr19_-_5142310 | 4.74 |
ENSDART00000130062
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr5_-_41672394 | 4.72 |
ENSDART00000164363
|
si:ch211-207c6.2
|
si:ch211-207c6.2 |
| chr3_+_48362748 | 4.71 |
ENSDART00000157199
|
mkl2b
|
MKL/myocardin-like 2b |
| chr1_+_45503061 | 4.63 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr21_-_11561855 | 4.63 |
ENSDART00000162426
|
cast
|
calpastatin |
| chr19_+_14197118 | 4.61 |
ENSDART00000166230
|
tpbga
|
trophoblast glycoprotein a |
| chr15_-_16076874 | 4.59 |
ENSDART00000144138
|
hnf1ba
|
HNF1 homeobox Ba |
| chr6_-_32106882 | 4.55 |
ENSDART00000144772
|
BX664614.1
|
ENSDARG00000095311 |
| chr1_+_41523935 | 4.55 |
ENSDART00000110860
|
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr25_+_13580892 | 4.53 |
ENSDART00000007029
|
usb1
|
U6 snRNA biogenesis 1 |
| chr6_-_49674729 | 4.51 |
ENSDART00000112226
|
apcdd1l
|
adenomatosis polyposis coli down-regulated 1-like |
| chr25_+_34135377 | 4.50 |
ENSDART00000157519
|
trpm1b
|
transient receptor potential cation channel, subfamily M, member 1b |
| chr20_+_40247918 | 4.42 |
ENSDART00000121818
|
trdn
|
triadin |
| chr12_-_34621359 | 4.42 |
|
|
|
| chr5_+_59845054 | 4.39 |
ENSDART00000130565
|
tmem132e
|
transmembrane protein 132E |
| chr7_+_56375651 | 4.36 |
ENSDART00000112242
|
zgc:194679
|
zgc:194679 |
| chr5_-_24568752 | 4.35 |
ENSDART00000145061
|
pnpla7b
|
patatin-like phospholipase domain containing 7b |
| chr1_-_46047321 | 4.35 |
|
|
|
| chr14_+_7626822 | 4.29 |
ENSDART00000109941
|
cxxc5b
|
CXXC finger protein 5b |
| chr16_-_10089440 | 4.29 |
ENSDART00000066372
|
id4
|
inhibitor of DNA binding 4 |
| chr17_+_24668907 | 4.25 |
ENSDART00000034263
ENSDART00000135794 |
sepn1
|
selenoprotein N, 1 |
| chr3_-_39287733 | 4.23 |
|
|
|
| chr25_-_13456748 | 4.22 |
ENSDART00000139290
|
ano10b
|
anoctamin 10b |
| chr12_-_36565562 | 4.21 |
ENSDART00000153259
|
si:ch211-216b21.2
|
si:ch211-216b21.2 |
| chr2_-_49114158 | 4.21 |
|
|
|
| chr10_-_27261937 | 4.18 |
|
|
|
| chr13_+_1741432 | 4.17 |
ENSDART00000161162
|
bmp5
|
bone morphogenetic protein 5 |
| chr5_+_59844830 | 4.16 |
ENSDART00000130565
|
tmem132e
|
transmembrane protein 132E |
| chr19_+_34582100 | 4.16 |
ENSDART00000135592
|
poc1bl
|
POC1 centriolar protein homolog B (Chlamydomonas), like |
| chr19_-_30975279 | 4.16 |
ENSDART00000171006
|
hpcal4
|
hippocalcin like 4 |
| chr14_+_49921861 | 4.14 |
ENSDART00000173240
|
zgc:154054
|
zgc:154054 |
| chr19_-_22957035 | 4.08 |
ENSDART00000019505
|
pleca
|
plectin a |
| chr12_+_28252623 | 4.08 |
ENSDART00000066294
|
cdk5r1b
|
cyclin-dependent kinase 5, regulatory subunit 1b (p35) |
| chr19_-_42721857 | 4.06 |
ENSDART00000150919
ENSDART00000151034 |
si:ch211-191i18.2
|
si:ch211-191i18.2 |
| chr6_-_43806125 | 4.06 |
|
|
|
| chr10_+_10393377 | 4.05 |
ENSDART00000109432
|
cercam
|
cerebral endothelial cell adhesion molecule |
| chr5_-_70939686 | 4.03 |
|
|
|
| chr13_+_17541579 | 4.03 |
ENSDART00000137776
|
comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr8_-_31044627 | 4.01 |
ENSDART00000109885
|
snrnp200
|
small nuclear ribonucleoprotein 200 (U5) |
| chr23_-_18203680 | 3.96 |
ENSDART00000016976
|
nucks1b
|
nuclear casein kinase and cyclin-dependent kinase substrate 1b |
| chr20_+_27194162 | 3.96 |
ENSDART00000024595
|
ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 |
| chr19_+_20202737 | 3.94 |
ENSDART00000164677
|
hoxa4a
|
homeobox A4a |
| chr13_+_1741474 | 3.93 |
ENSDART00000161162
|
bmp5
|
bone morphogenetic protein 5 |
| chr19_-_10635235 | 3.86 |
ENSDART00000104539
|
lim2.4
|
lens intrinsic membrane protein 2.4 |
| chr17_+_38314814 | 3.86 |
ENSDART00000017493
|
nkx2.1
|
NK2 homeobox 1 |
| chr25_-_13057808 | 3.84 |
ENSDART00000172571
|
smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr12_+_27026112 | 3.83 |
ENSDART00000076154
|
hoxb8b
|
homeobox B8b |
| chr12_-_7951265 | 3.82 |
|
|
|
| chr8_+_6989445 | 3.82 |
ENSDART00000134440
|
gpd1a
|
glycerol-3-phosphate dehydrogenase 1a |
| chr20_-_8036478 | 3.81 |
ENSDART00000083898
|
plpp3
|
phospholipid phosphatase 3 |
| chr10_+_17494274 | 3.80 |
|
|
|
| chr7_-_33558771 | 3.78 |
ENSDART00000074729
|
uacab
|
uveal autoantigen with coiled-coil domains and ankyrin repeats b |
| chr9_-_1969197 | 3.77 |
ENSDART00000080608
|
hoxd10a
|
homeobox D10a |
| chr19_+_23414927 | 3.73 |
ENSDART00000151090
|
gdf6b
|
growth differentiation factor 6b |
| chr9_+_8920445 | 3.69 |
ENSDART00000147820
|
carkd
|
carbohydrate kinase domain containing |
| chr5_-_57016269 | 3.68 |
ENSDART00000074264
|
crfb12
|
cytokine receptor family member B12 |
| chr6_-_54103765 | 3.64 |
ENSDART00000083880
|
hyal2a
|
hyaluronoglucosaminidase 2a |
| chr2_+_29862841 | 3.62 |
ENSDART00000135918
|
si:ch211-207d6.2
|
si:ch211-207d6.2 |
| chr3_-_58488929 | 3.59 |
ENSDART00000042386
|
unm_sa1261
|
un-named sa1261 |
| chr4_-_16417703 | 3.58 |
ENSDART00000013085
|
dcn
|
decorin |
| chr10_+_17893555 | 3.57 |
ENSDART00000113666
|
phf24
|
PHD finger protein 24 |
| chr16_-_13733759 | 3.57 |
ENSDART00000164344
|
si:dkeyp-69b9.6
|
si:dkeyp-69b9.6 |
| chr6_-_44282665 | 3.56 |
ENSDART00000157215
|
pdzrn3b
|
PDZ domain containing RING finger 3b |
| chr24_+_24924379 | 3.54 |
ENSDART00000115165
|
amer2
|
APC membrane recruitment protein 2 |
| chr1_-_24486146 | 3.53 |
ENSDART00000144711
|
tmem154
|
transmembrane protein 154 |
| chr14_-_1081316 | 3.53 |
|
|
|
| chr9_-_1983772 | 3.52 |
ENSDART00000082339
|
hoxd12a
|
homeobox D12a |
| chr12_+_2988101 | 3.52 |
ENSDART00000044690
ENSDART00000122905 |
rac3b
|
ras-related C3 botulinum toxin substrate 3b (rho family, small GTP binding protein Rac3) |
| chr19_-_5452918 | 3.50 |
ENSDART00000105004
|
krt17
|
keratin 17 |
| chr16_-_19154920 | 3.49 |
ENSDART00000088818
|
fhod3b
|
formin homology 2 domain containing 3b |
| chr13_+_23152038 | 3.49 |
ENSDART00000171676
|
khdrbs2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr24_+_19374200 | 3.48 |
ENSDART00000056081
ENSDART00000027022 |
sulf1
|
sulfatase 1 |
| chr21_-_24552672 | 3.44 |
|
|
|
| chr5_+_44722544 | 3.43 |
ENSDART00000084411
|
adamts3
|
ADAM metallopeptidase with thrombospondin type 1 motif, 3 |
| chr23_+_11350727 | 3.41 |
|
|
|
| chr16_+_1073570 | 3.40 |
|
|
|
| chr8_-_39944817 | 3.39 |
ENSDART00000083066
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr13_-_49695322 | 3.37 |
|
|
|
| chr19_-_22182031 | 3.36 |
ENSDART00000104279
|
znf516
|
zinc finger protein 516 |
| chr17_+_38307512 | 3.35 |
ENSDART00000005296
|
nkx2.9
|
NK2 transcription factor related, locus 9 (Drosophila) |
| chr8_+_39525254 | 3.33 |
|
|
|
| chr3_-_23513177 | 3.32 |
ENSDART00000078425
|
eve1
|
even-skipped-like1 |
| chr1_-_13547500 | 3.30 |
ENSDART00000044896
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr17_-_44133941 | 3.28 |
ENSDART00000126097
|
otx2
|
orthodenticle homeobox 2 |
| chr11_+_19894772 | 3.27 |
ENSDART00000103997
|
fezf2
|
FEZ family zinc finger 2 |
| chr3_-_12818954 | 3.26 |
ENSDART00000158747
ENSDART00000158815 |
pdgfab
|
platelet-derived growth factor alpha polypeptide b |
| chr11_-_3968366 | 3.25 |
ENSDART00000171093
|
glt8d1
|
glycosyltransferase 8 domain containing 1 |
| chr11_+_19894390 | 3.21 |
ENSDART00000103997
|
fezf2
|
FEZ family zinc finger 2 |
| chr9_-_47042015 | 3.20 |
ENSDART00000054137
|
igfbp5b
|
insulin-like growth factor binding protein 5b |
| chr23_+_44782492 | 3.16 |
ENSDART00000125198
|
pfn1
|
profilin 1 |
| chr14_+_7145893 | 3.15 |
ENSDART00000130388
|
gfra3
|
GDNF family receptor alpha 3 |
| chr9_+_32267615 | 3.15 |
|
|
|
| chr1_+_14451504 | 3.12 |
ENSDART00000111475
|
pcdh7a
|
protocadherin 7a |
| chr11_-_27253835 | 3.10 |
ENSDART00000065889
|
wnt7aa
|
wingless-type MMTV integration site family, member 7Aa |
| chr23_-_4175790 | 3.09 |
ENSDART00000109807
|
ENSDARG00000076299
|
ENSDARG00000076299 |
| chr14_-_29552242 | 2.93 |
ENSDART00000088004
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr19_+_38033219 | 2.93 |
ENSDART00000158960
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr7_-_29300402 | 2.93 |
ENSDART00000099477
|
rorab
|
RAR-related orphan receptor A, paralog b |
| KN150196v1_+_15525 | 2.92 |
|
|
|
| chr19_+_42491631 | 2.91 |
ENSDART00000150949
|
nfyc
|
nuclear transcription factor Y, gamma |
| chr3_-_54414513 | 2.90 |
ENSDART00000053106
|
s1pr2
|
sphingosine-1-phosphate receptor 2 |
| chr19_+_21783429 | 2.90 |
ENSDART00000024639
|
tshz1
|
teashirt zinc finger homeobox 1 |
| chr12_-_10384063 | 2.89 |
ENSDART00000052004
|
zgc:153595
|
zgc:153595 |
| chr10_-_7827244 | 2.87 |
ENSDART00000111058
|
mpx
|
myeloid-specific peroxidase |
| chr17_-_29885237 | 2.86 |
ENSDART00000009104
|
esrrga
|
estrogen-related receptor gamma a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 10.6 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 2.6 | 13.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 2.4 | 7.3 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 2.4 | 14.1 | GO:0003311 | pancreatic D cell differentiation(GO:0003311) |
| 2.2 | 8.8 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 2.2 | 10.9 | GO:0021982 | pineal gland development(GO:0021982) |
| 1.9 | 7.7 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 1.8 | 5.5 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 1.4 | 8.7 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 1.4 | 5.6 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 1.3 | 9.3 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 1.3 | 5.3 | GO:1901052 | sarcosine metabolic process(GO:1901052) |
| 1.3 | 12.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 1.2 | 11.0 | GO:0060114 | vestibular receptor cell differentiation(GO:0060114) vestibular receptor cell development(GO:0060118) |
| 1.2 | 2.4 | GO:1901490 | regulation of lymphangiogenesis(GO:1901490) positive regulation of lymphangiogenesis(GO:1901492) |
| 1.2 | 7.0 | GO:0071326 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 1.1 | 4.6 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 1.1 | 6.6 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 1.1 | 5.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 1.0 | 9.4 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 1.0 | 2.9 | GO:0035676 | anterior lateral line neuromast hair cell development(GO:0035676) |
| 0.9 | 4.7 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.9 | 5.5 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.9 | 2.7 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.9 | 12.3 | GO:0036302 | atrioventricular canal development(GO:0036302) |
| 0.9 | 3.5 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.8 | 4.1 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.8 | 4.9 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.8 | 2.4 | GO:0031639 | regulation of collagen metabolic process(GO:0010712) regulation of granulocyte differentiation(GO:0030852) plasminogen activation(GO:0031639) regulation of collagen biosynthetic process(GO:0032965) regulation of multicellular organismal metabolic process(GO:0044246) regulation of neutrophil differentiation(GO:0045658) |
| 0.8 | 7.8 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.8 | 7.7 | GO:0048730 | epidermis morphogenesis(GO:0048730) |
| 0.8 | 3.8 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.8 | 5.3 | GO:0036065 | fucosylation(GO:0036065) |
| 0.7 | 4.5 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.7 | 1.5 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.7 | 3.4 | GO:0021572 | rhombomere 5 development(GO:0021571) rhombomere 6 development(GO:0021572) |
| 0.7 | 2.0 | GO:0061550 | cranial ganglion development(GO:0061550) |
| 0.7 | 10.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.6 | 9.0 | GO:0021654 | rhombomere boundary formation(GO:0021654) |
| 0.6 | 6.4 | GO:0042310 | vasoconstriction(GO:0042310) |
| 0.6 | 3.1 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.6 | 5.4 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.6 | 2.3 | GO:0045168 | cell-cell signaling involved in cell fate commitment(GO:0045168) |
| 0.6 | 1.7 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.5 | 1.5 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.5 | 5.5 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.5 | 2.4 | GO:0060579 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) ventral spinal cord interneuron fate commitment(GO:0060579) |
| 0.5 | 4.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.5 | 5.6 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.4 | 3.3 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.4 | 2.0 | GO:1902221 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.4 | 8.6 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.4 | 6.5 | GO:0014034 | neural crest cell fate commitment(GO:0014034) neural crest cell fate specification(GO:0014036) |
| 0.4 | 1.9 | GO:0021988 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.4 | 11.8 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.4 | 1.1 | GO:0000388 | spliceosome conformational change to release U4 (or U4atac) and U1 (or U11)(GO:0000388) |
| 0.4 | 4.0 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.4 | 8.6 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.3 | 1.4 | GO:0035776 | pronephric proximal tubule development(GO:0035776) |
| 0.3 | 1.7 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.3 | 1.9 | GO:0046822 | regulation of nucleocytoplasmic transport(GO:0046822) |
| 0.3 | 4.4 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.3 | 6.8 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.3 | 2.2 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.3 | 5.0 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.3 | 2.7 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.3 | 4.5 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.3 | 5.0 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.3 | 0.8 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.3 | 6.9 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.2 | 3.8 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.2 | 3.6 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.2 | 0.7 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.2 | 1.8 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.2 | 0.9 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.2 | 1.5 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.2 | 2.9 | GO:2000377 | regulation of reactive oxygen species metabolic process(GO:2000377) |
| 0.2 | 6.6 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.2 | 0.8 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.2 | 2.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.2 | 5.4 | GO:0000302 | response to reactive oxygen species(GO:0000302) |
| 0.2 | 1.8 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.2 | 4.5 | GO:0051262 | protein tetramerization(GO:0051262) |
| 0.2 | 2.5 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.2 | 1.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 2.6 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.2 | 1.4 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.2 | 1.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 2.5 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.2 | 4.1 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 5.5 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 3.1 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.1 | 4.0 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.1 | 6.6 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.1 | 1.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 4.9 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.1 | 4.8 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) actin filament capping(GO:0051693) |
| 0.1 | 9.3 | GO:0002521 | leukocyte differentiation(GO:0002521) |
| 0.1 | 2.7 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.1 | 1.3 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 1.7 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 4.1 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.1 | 5.9 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 10.1 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.1 | 6.6 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 0.7 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.1 | 3.7 | GO:0007599 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.1 | 0.6 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.1 | 2.8 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 7.6 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.1 | 2.9 | GO:0071599 | otic vesicle development(GO:0071599) |
| 0.1 | 2.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 8.4 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.1 | 9.0 | GO:0001947 | heart looping(GO:0001947) |
| 0.1 | 2.8 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.1 | 5.0 | GO:0008544 | epidermis development(GO:0008544) |
| 0.1 | 2.1 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 6.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 1.9 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.1 | 13.6 | GO:0000280 | nuclear division(GO:0000280) |
| 0.1 | 0.6 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.1 | 17.2 | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter(GO:0045944) |
| 0.0 | 4.1 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 5.7 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 2.7 | GO:0002088 | lens development in camera-type eye(GO:0002088) |
| 0.0 | 3.9 | GO:0030111 | regulation of Wnt signaling pathway(GO:0030111) |
| 0.0 | 0.9 | GO:0043652 | engulfment of apoptotic cell(GO:0043652) |
| 0.0 | 2.6 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 4.6 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 1.1 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 1.5 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.2 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 1.4 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) peptidyl-threonine modification(GO:0018210) |
| 0.0 | 0.4 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.0 | 3.4 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 3.8 | GO:0045664 | regulation of neuron differentiation(GO:0045664) |
| 0.0 | 0.3 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.6 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 3.1 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 1.4 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 1.3 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 1.1 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 26.0 | GO:0006357 | regulation of transcription from RNA polymerase II promoter(GO:0006357) |
| 0.0 | 1.4 | GO:0033674 | positive regulation of kinase activity(GO:0033674) |
| 0.0 | 1.2 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 1.0 | GO:0051169 | nucleocytoplasmic transport(GO:0006913) nuclear transport(GO:0051169) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 1.4 | 7.2 | GO:0008091 | spectrin(GO:0008091) |
| 1.0 | 10.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.6 | 3.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.6 | 42.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.5 | 1.6 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.5 | 5.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.4 | 2.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.3 | 11.0 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.3 | 2.9 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.3 | 2.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.2 | 2.9 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 4.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.2 | 14.3 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.2 | 1.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.2 | 5.5 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.2 | 2.7 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 0.7 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.1 | 0.6 | GO:0043034 | costamere(GO:0043034) |
| 0.1 | 7.7 | GO:0030018 | Z disc(GO:0030018) |
| 0.1 | 1.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 5.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 0.7 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 2.4 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 4.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 27.2 | GO:0000785 | chromatin(GO:0000785) |
| 0.1 | 13.3 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.1 | 46.6 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 20.2 | GO:0005576 | extracellular region(GO:0005576) |
| 0.1 | 3.6 | GO:0045202 | synapse(GO:0045202) |
| 0.1 | 3.1 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 1.6 | GO:0099501 | synaptic vesicle membrane(GO:0030672) exocytic vesicle membrane(GO:0099501) |
| 0.0 | 17.9 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 0.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 1.7 | GO:0005924 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 18.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 2.4 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 8.9 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 1.5 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 2.7 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.2 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 7.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 6.2 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.0 | 0.8 | GO:0016324 | apical plasma membrane(GO:0016324) apical part of cell(GO:0045177) |
| 0.0 | 125.4 | GO:0005634 | nucleus(GO:0005634) |
| 0.0 | 1.4 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 67.5 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.8 | GO:0005694 | chromosome(GO:0005694) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0016175 | NAD(P)H oxidase activity(GO:0016174) superoxide-generating NADPH oxidase activity(GO:0016175) |
| 2.1 | 14.4 | GO:0070888 | E-box binding(GO:0070888) |
| 1.8 | 5.3 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 1.7 | 7.0 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.6 | 14.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 1.6 | 4.7 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 1.4 | 7.0 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 1.4 | 6.9 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 1.3 | 13.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 1.3 | 12.7 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 1.0 | 4.0 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 1.0 | 4.9 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 1.0 | 10.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.9 | 8.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.8 | 4.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.8 | 5.5 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.8 | 3.8 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.7 | 2.9 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.7 | 2.6 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.6 | 5.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.6 | 6.1 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.6 | 6.0 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.6 | 2.4 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.6 | 5.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.6 | 10.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.5 | 4.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.5 | 4.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.4 | 1.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.4 | 3.3 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.4 | 5.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.4 | 2.7 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.4 | 6.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.3 | 1.7 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.3 | 4.3 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.3 | 2.2 | GO:0015145 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.3 | 3.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.3 | 4.4 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.3 | 3.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 2.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 3.1 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.3 | 1.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.3 | 4.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.2 | 4.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.2 | 4.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 3.9 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 0.7 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.2 | 9.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 2.4 | GO:0048018 | receptor activator activity(GO:0030546) receptor agonist activity(GO:0048018) |
| 0.2 | 10.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.2 | 5.9 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.2 | 7.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 8.0 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.2 | 1.5 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.2 | 9.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.2 | 4.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.2 | 3.3 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 6.6 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 2.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 2.9 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 1.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.1 | 3.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 2.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.1 | 4.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 5.5 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 4.5 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 1.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 29.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 6.6 | GO:0051287 | NAD binding(GO:0051287) |
| 0.1 | 1.8 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 2.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 3.7 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 1.6 | GO:0005539 | glycosaminoglycan binding(GO:0005539) |
| 0.1 | 5.1 | GO:0016859 | cis-trans isomerase activity(GO:0016859) |
| 0.1 | 39.4 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.1 | 0.9 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 5.5 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 4.1 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.1 | 4.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 118.0 | GO:0000981 | RNA polymerase II transcription factor activity, sequence-specific DNA binding(GO:0000981) |
| 0.1 | 3.7 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 3.5 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 2.5 | GO:0008194 | UDP-glycosyltransferase activity(GO:0008194) |
| 0.0 | 1.5 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 14.0 | GO:0003712 | transcription factor activity, transcription factor binding(GO:0000989) transcription cofactor activity(GO:0003712) |
| 0.0 | 3.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 3.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 6.7 | GO:0016757 | transferase activity, transferring glycosyl groups(GO:0016757) |
| 0.0 | 4.6 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 12.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.9 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 2.9 | GO:0003700 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 4.0 | GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity(GO:0008757) |
| 0.0 | 2.2 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 5.7 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.6 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.1 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 1.3 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.9 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 0.5 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.3 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0009931 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 18.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.5 | 8.3 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.4 | 14.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.4 | 6.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.4 | 10.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.3 | 2.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.3 | 2.9 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.2 | 2.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 11.0 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.2 | 9.2 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.1 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 3.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 9.5 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.1 | 2.4 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 6.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 2.2 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 1.8 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 1.7 | 13.6 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 1.2 | 2.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 1.0 | 7.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.7 | 14.2 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.7 | 14.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.5 | 16.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.5 | 8.6 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.4 | 2.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.4 | 2.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 2.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.2 | 10.6 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.2 | 1.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.2 | 1.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 1.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.2 | 2.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.2 | 2.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 1.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 3.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.1 | 2.9 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.1 | 1.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 3.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 1.8 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 0.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 1.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 2.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.1 | 5.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 0.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.8 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.6 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 1.1 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.7 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 1.0 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |