Project
DANIO-CODE
Navigation
Downloads
Results for si:ch211-153j24.3
Z-value: 2.56
Transcription factors associated with si:ch211-153j24.3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
si_ch211-153j24.3
|
ENSDARG00000068428 | si_ch211-153j24.3 |
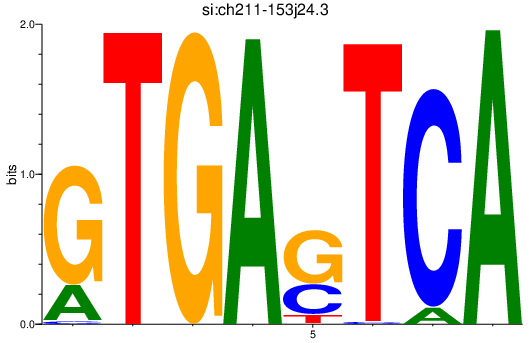
Activity profile of si:ch211-153j24.3 motif
Sorted Z-values of si:ch211-153j24.3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of si:ch211-153j24.3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_37934447 | 9.70 |
ENSDART00000076082
|
fetub
|
fetuin B |
| chr23_-_1008307 | 8.70 |
ENSDART00000110588
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr9_-_48673183 | 8.29 |
ENSDART00000140185
ENSDART00000134185 |
col28a2a
|
collagen, type XXVIII, alpha 2a |
| chr20_+_26981663 | 7.60 |
ENSDART00000077769
|
serpinb1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1 |
| chr20_-_26632676 | 7.55 |
ENSDART00000131994
|
mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr16_+_21109486 | 7.11 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr22_-_15567180 | 6.96 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr3_+_24067387 | 6.50 |
ENSDART00000055609
|
atf4b
|
activating transcription factor 4b |
| chr5_-_66792947 | 6.18 |
ENSDART00000147009
|
si:dkey-251i10.2
|
si:dkey-251i10.2 |
| chr1_+_41148042 | 6.12 |
ENSDART00000145170
ENSDART00000136879 |
smox
|
spermine oxidase |
| chr13_-_36996246 | 6.11 |
ENSDART00000133242
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr25_+_22489716 | 6.04 |
ENSDART00000127831
|
stra6
|
stimulated by retinoic acid 6 |
| chr7_+_44373815 | 6.03 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr3_+_26896869 | 6.02 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr25_+_18487408 | 5.98 |
ENSDART00000148741
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr1_-_5048203 | 5.88 |
ENSDART00000150863
ENSDART00000163417 |
nrp2a
|
neuropilin 2a |
| chr1_-_9387290 | 5.86 |
ENSDART00000135522
ENSDART00000135676 |
fga
|
fibrinogen alpha chain |
| chr11_+_26371444 | 5.76 |
ENSDART00000042322
|
map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr10_+_35535209 | 5.71 |
ENSDART00000109705
|
phldb2a
|
pleckstrin homology-like domain, family B, member 2a |
| chr4_-_15442828 | 5.71 |
ENSDART00000157414
|
plxna4
|
plexin A4 |
| chr14_-_2008649 | 5.45 |
ENSDART00000161817
|
pcdh2g16
|
protocadherin 2 gamma 16 |
| chr9_-_11589126 | 5.40 |
ENSDART00000146832
|
cryba2b
|
crystallin, beta A2b |
| chr16_+_33702010 | 5.25 |
ENSDART00000143757
|
fhl3a
|
four and a half LIM domains 3a |
| chr21_+_22808694 | 5.20 |
ENSDART00000065555
|
birc2
|
baculoviral IAP repeat containing 2 |
| chr15_+_9351511 | 5.18 |
ENSDART00000144381
|
sgcg
|
sarcoglycan, gamma |
| chr19_-_23037220 | 5.14 |
ENSDART00000090669
|
pleca
|
plectin a |
| chr13_+_46899292 | 5.10 |
|
|
|
| chr5_+_15319430 | 5.06 |
ENSDART00000162003
|
hspb8
|
heat shock protein b8 |
| chr3_-_16056593 | 5.01 |
|
|
|
| chr6_+_54703206 | 4.99 |
ENSDART00000074605
|
pkp1b
|
plakophilin 1b |
| chr10_+_16111842 | 4.98 |
ENSDART00000141654
|
megf10
|
multiple EGF-like-domains 10 |
| chr4_+_1750689 | 4.94 |
ENSDART00000146779
|
slc38a2
|
solute carrier family 38, member 2 |
| chr25_-_18234069 | 4.82 |
ENSDART00000104496
|
dusp6
|
dual specificity phosphatase 6 |
| chr1_-_22170553 | 4.81 |
ENSDART00000139412
|
SMIM18
|
small integral membrane protein 18 |
| chr21_+_25729090 | 4.78 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr25_-_22089794 | 4.77 |
ENSDART00000139110
|
pkp3a
|
plakophilin 3a |
| chr7_+_35769973 | 4.69 |
ENSDART00000168658
|
irx3a
|
iroquois homeobox 3a |
| KN150349v1_-_13313 | 4.51 |
|
|
|
| chr6_+_29800606 | 4.47 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr5_+_17120453 | 4.46 |
|
|
|
| chr17_+_25503946 | 4.37 |
|
|
|
| chr24_+_24308055 | 4.36 |
|
|
|
| chr10_+_32007448 | 4.30 |
ENSDART00000019416
|
lhfp
|
lipoma HMGIC fusion partner |
| chr11_+_37639045 | 4.30 |
ENSDART00000111157
|
si:ch211-112f3.4
|
si:ch211-112f3.4 |
| chr11_-_13069266 | 4.27 |
ENSDART00000169052
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr22_+_16471319 | 4.24 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr19_+_17451381 | 4.23 |
ENSDART00000167602
|
spaca4l
|
sperm acrosome associated 4 like |
| chr13_+_23065500 | 4.19 |
ENSDART00000158370
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr3_+_16826320 | 4.18 |
ENSDART00000112450
|
cavin1a
|
caveolae associated protein 1a |
| chr6_-_13654186 | 4.06 |
ENSDART00000150102
ENSDART00000041269 |
cryba2a
|
crystallin, beta A2a |
| chr1_-_416138 | 4.05 |
ENSDART00000092524
|
rasa3
|
RAS p21 protein activator 3 |
| chr2_+_47727856 | 4.04 |
ENSDART00000112579
|
scg2b
|
secretogranin II (chromogranin C), b |
| chr12_-_1931281 | 4.00 |
ENSDART00000005676
ENSDART00000127937 |
sox9a
|
SRY (sex determining region Y)-box 9a |
| chr19_-_6466494 | 3.98 |
ENSDART00000104950
|
atp1a3a
|
ATPase, Na+/K+ transporting, alpha 3a polypeptide |
| chr5_-_65349550 | 3.97 |
ENSDART00000164228
|
nrarpb
|
notch-regulated ankyrin repeat protein b |
| chr11_-_6004509 | 3.93 |
ENSDART00000108628
|
ano8b
|
anoctamin 8b |
| chr14_-_41311458 | 3.92 |
ENSDART00000163039
|
fgfrl1b
|
fibroblast growth factor receptor-like 1b |
| chr9_+_25964943 | 3.88 |
ENSDART00000147229
ENSDART00000127834 |
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr16_+_42925950 | 3.88 |
ENSDART00000159730
ENSDART00000014956 |
polr3glb
|
polymerase (RNA) III (DNA directed) polypeptide G like b |
| chr11_-_23151247 | 3.85 |
|
|
|
| chr12_-_4648262 | 3.84 |
ENSDART00000152771
|
si:ch211-255p10.3
|
si:ch211-255p10.3 |
| chr7_+_38479571 | 3.83 |
ENSDART00000170486
|
f2
|
coagulation factor II (thrombin) |
| chr7_-_28425307 | 3.83 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr7_+_20046425 | 3.79 |
ENSDART00000131019
|
acadvl
|
acyl-CoA dehydrogenase, very long chain |
| chr6_+_2768475 | 3.72 |
|
|
|
| chr14_-_26138828 | 3.72 |
ENSDART00000140173
|
si:dkeyp-110e4.6
|
si:dkeyp-110e4.6 |
| chr5_+_42312784 | 3.65 |
ENSDART00000039973
|
rufy3
|
RUN and FYVE domain containing 3 |
| chr25_-_18044103 | 3.64 |
ENSDART00000113581
|
kitlga
|
kit ligand a |
| chr14_+_34146377 | 3.63 |
ENSDART00000131861
|
tmsb2
|
thymosin beta 2 |
| chr14_-_40454194 | 3.61 |
ENSDART00000166621
|
ef1
|
E74-like factor 1 (ets domain transcription factor) |
| chr24_+_26922969 | 3.59 |
|
|
|
| chr16_-_21981065 | 3.58 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr9_-_35106146 | 3.55 |
ENSDART00000139608
ENSDART00000100728 ENSDART00000123005 |
upf3a
|
UPF3 regulator of nonsense transcripts homolog A (yeast) |
| chr24_+_36478896 | 3.54 |
|
|
|
| chr20_-_13728005 | 3.51 |
ENSDART00000152499
|
ezrb
|
ezrin b |
| chr9_-_47957181 | 3.47 |
ENSDART00000015159
|
tns1b
|
tensin 1b |
| chr10_+_35478939 | 3.46 |
ENSDART00000147303
|
hhla2a.1
|
HERV-H LTR-associating 2a, tandem duplicate 1 |
| chr3_-_39245184 | 3.46 |
|
|
|
| chr14_-_15651090 | 3.42 |
ENSDART00000169197
|
flt4
|
fms-related tyrosine kinase 4 |
| chr23_+_35819625 | 3.40 |
ENSDART00000049551
|
rarga
|
retinoic acid receptor gamma a |
| chr20_-_39200252 | 3.38 |
ENSDART00000037318
ENSDART00000143379 |
rcan2
|
regulator of calcineurin 2 |
| chr16_+_33701759 | 3.35 |
ENSDART00000143757
|
fhl3a
|
four and a half LIM domains 3a |
| chr25_+_18487313 | 3.35 |
ENSDART00000148741
|
met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr6_+_19795100 | 3.30 |
|
|
|
| chr17_+_45430353 | 3.23 |
ENSDART00000162937
|
ezra
|
ezrin a |
| chr24_+_20430778 | 3.22 |
ENSDART00000010488
|
klhl40b
|
kelch-like family member 40b |
| chr15_+_29056721 | 3.21 |
ENSDART00000076648
|
clip3
|
CAP-GLY domain containing linker protein 3 |
| KN150583v1_-_1389 | 3.20 |
|
|
|
| chr21_-_18787657 | 3.17 |
|
|
|
| chr1_-_22144014 | 3.17 |
ENSDART00000043556
|
ldb2b
|
LIM domain binding 2b |
| chr15_-_19836573 | 3.17 |
ENSDART00000114888
|
picalmb
|
phosphatidylinositol binding clathrin assembly protein b |
| chr5_-_3489302 | 3.11 |
|
|
|
| chr6_-_18733424 | 3.07 |
ENSDART00000151578
|
tns1a
|
tensin 1a |
| chr7_+_24762755 | 3.05 |
ENSDART00000170873
|
sb:cb1058
|
sb:cb1058 |
| chr8_-_38168395 | 3.04 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr13_-_7897936 | 3.03 |
ENSDART00000139728
|
si:ch211-250c4.4
|
si:ch211-250c4.4 |
| chr6_+_54931721 | 3.01 |
|
|
|
| chr21_-_44570264 | 3.00 |
ENSDART00000159323
|
fundc2
|
fun14 domain containing 2 |
| chr21_+_30757831 | 2.99 |
ENSDART00000139486
|
ENSDARG00000030006
|
ENSDARG00000030006 |
| chr20_-_23539928 | 2.98 |
ENSDART00000022887
|
slc10a4
|
solute carrier family 10, member 4 |
| chr25_-_22890657 | 2.98 |
|
|
|
| chr18_-_8355104 | 2.95 |
ENSDART00000148243
|
mapk8ip2
|
mitogen-activated protein kinase 8 interacting protein 2 |
| chr5_+_51992974 | 2.90 |
ENSDART00000170341
|
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr5_-_39696678 | 2.90 |
ENSDART00000097526
|
hspb3
|
heat shock protein, alpha-crystallin-related, b3 |
| chr19_-_33624795 | 2.89 |
ENSDART00000109868
|
trib1
|
tribbles pseudokinase 1 |
| chr3_+_25933164 | 2.88 |
ENSDART00000143697
|
si:dkeyp-69e1.8
|
si:dkeyp-69e1.8 |
| chr23_-_28367816 | 2.88 |
ENSDART00000003548
|
znf385a
|
zinc finger protein 385A |
| chr1_-_51862897 | 2.86 |
ENSDART00000136469
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr7_-_25426346 | 2.85 |
ENSDART00000082620
|
dysf
|
dysferlin, limb girdle muscular dystrophy 2B (autosomal recessive) |
| chr16_-_34304851 | 2.85 |
ENSDART00000145485
|
phactr4b
|
phosphatase and actin regulator 4b |
| chr9_-_12726136 | 2.77 |
|
|
|
| chr21_-_8304754 | 2.73 |
ENSDART00000055328
|
nek6
|
NIMA-related kinase 6 |
| chr1_-_44199728 | 2.72 |
ENSDART00000145354
|
tcirg1a
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3a |
| chr23_-_1008468 | 2.68 |
ENSDART00000110588
|
cdh26.1
|
cadherin 26, tandem duplicate 1 |
| chr10_+_9764591 | 2.67 |
ENSDART00000091780
|
rc3h2
|
ring finger and CCCH-type domains 2 |
| chr19_-_31448000 | 2.67 |
ENSDART00000147504
|
bzw2
|
basic leucine zipper and W2 domains 2 |
| chr11_+_18020191 | 2.66 |
ENSDART00000125984
|
mustn1b
|
musculoskeletal, embryonic nuclear protein 1b |
| chr22_-_29387056 | 2.66 |
ENSDART00000121599
|
pdgfba
|
platelet-derived growth factor beta polypeptide a |
| chr6_+_13099562 | 2.65 |
ENSDART00000038505
|
rprmb
|
reprimo, TP53 dependent G2 arrest mediator candidate b |
| chr2_-_42011586 | 2.63 |
ENSDART00000045763
|
keap1a
|
kelch-like ECH-associated protein 1a |
| chr16_+_11138924 | 2.62 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr19_-_7576069 | 2.60 |
ENSDART00000148836
|
rfx5
|
regulatory factor X, 5 |
| chr12_-_28248133 | 2.59 |
ENSDART00000016283
|
psmd11b
|
proteasome 26S subunit, non-ATPase 11b |
| chr25_-_13737344 | 2.58 |
|
|
|
| chr17_+_51272621 | 2.55 |
|
|
|
| chr20_-_25726868 | 2.55 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr5_-_43121949 | 2.53 |
ENSDART00000157093
|
si:dkey-40c11.1
|
si:dkey-40c11.1 |
| KN149726v1_+_1094 | 2.53 |
|
|
|
| chr5_-_29891016 | 2.51 |
ENSDART00000138464
|
phldb1a
|
pleckstrin homology-like domain, family B, member 1a |
| chr16_+_11138879 | 2.50 |
ENSDART00000091183
|
erfl3
|
Ets2 repressor factor like 3 |
| chr16_+_13928376 | 2.47 |
ENSDART00000163251
|
flcn
|
folliculin |
| chr15_-_40391882 | 2.46 |
|
|
|
| chr16_+_13928844 | 2.46 |
ENSDART00000090191
|
flcn
|
folliculin |
| chr20_+_30948175 | 2.46 |
|
|
|
| chr16_-_33105847 | 2.43 |
|
|
|
| chr8_-_39944817 | 2.42 |
ENSDART00000083066
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr21_+_1504942 | 2.40 |
ENSDART00000130274
|
rab27b
|
RAB27B, member RAS oncogene family |
| chr20_+_19175518 | 2.39 |
|
|
|
| chr11_-_11925832 | 2.38 |
|
|
|
| chr20_+_27194162 | 2.38 |
ENSDART00000024595
|
ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 |
| chr1_+_16983775 | 2.37 |
ENSDART00000030665
|
slc25a4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr7_+_60992913 | 2.35 |
ENSDART00000083255
|
adam19a
|
ADAM metallopeptidase domain 19a |
| chr7_+_23604092 | 2.35 |
ENSDART00000101406
|
rab39bb
|
RAB39B, member RAS oncogene family b |
| chr21_+_5597476 | 2.34 |
ENSDART00000161235
ENSDART00000170456 |
shroom3
|
shroom family member 3 |
| chr10_-_20567013 | 2.31 |
ENSDART00000159060
|
ddhd2
|
DDHD domain containing 2 |
| chr17_-_5426195 | 2.25 |
ENSDART00000035944
|
clic5a
|
chloride intracellular channel 5a |
| chr6_-_43030859 | 2.24 |
ENSDART00000161722
|
glyctk
|
glycerate kinase |
| chr10_+_4476558 | 2.24 |
ENSDART00000125299
|
plk2a
|
polo-like kinase 2a (Drosophila) |
| chr9_+_55778669 | 2.24 |
|
|
|
| chr13_+_23539260 | 2.23 |
ENSDART00000134973
|
pcnxl2
|
pecanex-like 2 (Drosophila) |
| chr8_+_25984588 | 2.22 |
|
|
|
| chr7_+_20046613 | 2.22 |
ENSDART00000131019
|
acadvl
|
acyl-CoA dehydrogenase, very long chain |
| chr5_+_60871396 | 2.19 |
ENSDART00000131937
|
orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr7_+_10346939 | 2.16 |
|
|
|
| chr3_-_45669791 | 2.16 |
ENSDART00000169364
|
gcgra
|
glucagon receptor a |
| chr19_+_5156446 | 2.15 |
ENSDART00000151681
|
eno2
|
enolase 2 |
| chr9_-_1964814 | 2.13 |
ENSDART00000082354
|
hoxd9a
|
homeobox D9a |
| chr19_+_15538967 | 2.12 |
ENSDART00000171403
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr2_+_25622497 | 2.12 |
ENSDART00000131977
|
ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr12_-_37124892 | 2.12 |
ENSDART00000146142
|
pmp22b
|
peripheral myelin protein 22b |
| chr3_+_23621843 | 2.11 |
ENSDART00000146636
|
hoxb2a
|
homeobox B2a |
| chr3_+_28450576 | 2.09 |
ENSDART00000150893
|
sept12
|
septin 12 |
| chr18_-_17031173 | 2.07 |
ENSDART00000129146
|
tbc1d15
|
TBC1 domain family, member 15 |
| chr21_-_35646193 | 2.04 |
|
|
|
| chr17_-_35011682 | 2.02 |
|
|
|
| chr3_+_20007257 | 2.02 |
ENSDART00000125281
|
ngfra
|
nerve growth factor receptor a (TNFR superfamily, member 16) |
| chr6_-_39313219 | 2.01 |
|
|
|
| chr10_+_1610794 | 1.98 |
ENSDART00000060946
|
sgsm1b
|
small G protein signaling modulator 1b |
| chr17_+_45832040 | 1.97 |
ENSDART00000154638
|
kif26ab
|
kinesin family member 26Ab |
| chr22_-_864745 | 1.96 |
ENSDART00000035514
|
cept1b
|
choline/ethanolamine phosphotransferase 1b |
| chr18_+_27205666 | 1.96 |
|
|
|
| chr2_+_19587617 | 1.95 |
ENSDART00000166292
|
cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr7_-_39266252 | 1.94 |
ENSDART00000173965
|
otog
|
otogelin |
| chr4_-_26262761 | 1.94 |
ENSDART00000176623
|
BX957270.2
|
ENSDARG00000107325 |
| chr21_-_30508374 | 1.93 |
ENSDART00000019199
|
rab39ba
|
RAB39B, member RAS oncogene family a |
| chr19_-_37087393 | 1.92 |
ENSDART00000001805
|
csmd2
|
CUB and Sushi multiple domains 2 |
| chr5_+_69401770 | 1.92 |
|
|
|
| chr3_-_12087518 | 1.91 |
ENSDART00000081374
|
cfap70
|
cilia and flagella associated protein 70 |
| chr14_-_33704021 | 1.91 |
ENSDART00000149396
ENSDART00000123607 |
cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr21_+_9483423 | 1.89 |
ENSDART00000162834
|
mapk10
|
mitogen-activated protein kinase 10 |
| chr19_-_41819752 | 1.89 |
ENSDART00000167772
|
shfm1
|
split hand/foot malformation (ectrodactyly) type 1 |
| chr14_-_17282615 | 1.88 |
ENSDART00000006716
ENSDART00000136242 |
selt2
|
selenoprotein T, 2 |
| chr9_+_25964868 | 1.88 |
ENSDART00000147229
ENSDART00000127834 |
zeb2a
|
zinc finger E-box binding homeobox 2a |
| chr16_-_43161408 | 1.87 |
ENSDART00000149431
|
sema4aa
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Aa |
| chr22_+_16279764 | 1.86 |
|
|
|
| chr17_-_49931552 | 1.86 |
ENSDART00000161008
|
filip1a
|
filamin A interacting protein 1a |
| chr14_+_31564252 | 1.86 |
|
|
|
| chr5_-_13318226 | 1.85 |
|
|
|
| chr3_-_16056497 | 1.85 |
|
|
|
| chr22_+_37696217 | 1.84 |
ENSDART00000007346
|
psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr7_-_18448121 | 1.84 |
ENSDART00000112359
|
si:ch211-119e14.9
|
si:ch211-119e14.9 |
| chr1_-_47080507 | 1.82 |
ENSDART00000101079
|
neurl1aa
|
neuralized E3 ubiquitin protein ligase 1Aa |
| chr5_+_23585492 | 1.80 |
ENSDART00000135083
|
tp53
|
tumor protein p53 |
| chr4_-_28384562 | 1.79 |
ENSDART00000056132
|
tprkb
|
Tp53rk binding protein |
| chr22_+_4563528 | 1.78 |
ENSDART00000122324
|
si:ch1073-104i17.1
|
si:ch1073-104i17.1 |
| chr23_-_5429419 | 1.78 |
ENSDART00000159105
|
CR936540.3
|
ENSDARG00000101435 |
| chr22_+_35113233 | 1.77 |
ENSDART00000123066
|
srfa
|
serum response factor a |
| chr6_-_39313420 | 1.74 |
|
|
|
| chr7_-_68033217 | 1.74 |
|
|
|
| chr11_+_26152136 | 1.74 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:1905178 | regulation of cardiac muscle tissue regeneration(GO:1905178) |
| 1.3 | 5.2 | GO:0010939 | regulation of necrotic cell death(GO:0010939) regulation of necroptotic process(GO:0060544) |
| 1.3 | 3.8 | GO:0070445 | peripheral nervous system myelin maintenance(GO:0032287) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 1.2 | 3.5 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 1.1 | 3.4 | GO:0035474 | selective angioblast sprouting(GO:0035474) venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 1.1 | 3.2 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 1.0 | 6.9 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 1.0 | 5.8 | GO:0090243 | fibroblast growth factor receptor signaling pathway involved in somitogenesis(GO:0090243) |
| 1.0 | 3.8 | GO:0050820 | positive regulation of blood coagulation(GO:0030194) positive regulation of coagulation(GO:0050820) positive regulation of hemostasis(GO:1900048) |
| 0.9 | 2.8 | GO:0050765 | monocyte activation involved in immune response(GO:0002280) negative regulation of phagocytosis(GO:0050765) |
| 0.9 | 9.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.9 | 6.0 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.8 | 9.3 | GO:0021683 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.8 | 5.0 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.8 | 7.5 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.8 | 6.0 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.7 | 4.9 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.7 | 2.8 | GO:0015860 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.6 | 1.9 | GO:0032475 | otolith formation(GO:0032475) |
| 0.6 | 2.4 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.6 | 3.6 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.6 | 4.2 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 0.6 | 1.8 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.6 | 4.0 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.5 | 2.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.5 | 5.9 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.5 | 2.7 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.5 | 2.6 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.5 | 1.6 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.5 | 4.2 | GO:0032185 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.5 | 3.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.5 | 4.9 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) positive regulation of TORC1 signaling(GO:1904263) |
| 0.4 | 0.9 | GO:0007613 | memory(GO:0007613) |
| 0.4 | 1.2 | GO:0097104 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.4 | 1.6 | GO:0046100 | guanine salvage(GO:0006178) GMP salvage(GO:0032263) guanine metabolic process(GO:0046098) guanine biosynthetic process(GO:0046099) hypoxanthine metabolic process(GO:0046100) |
| 0.4 | 5.8 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.4 | 17.3 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.4 | 1.1 | GO:0072672 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.4 | 3.2 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.4 | 1.1 | GO:1905207 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.3 | 2.4 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.3 | 1.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.3 | 2.7 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.3 | 4.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.3 | 2.0 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.3 | 5.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 0.9 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.3 | 4.0 | GO:0043363 | nucleate erythrocyte differentiation(GO:0043363) |
| 0.3 | 5.9 | GO:0038084 | vascular endothelial growth factor signaling pathway(GO:0038084) |
| 0.3 | 4.0 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.3 | 7.0 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.3 | 4.3 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.2 | 2.7 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.2 | 1.7 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.2 | 1.9 | GO:0001964 | startle response(GO:0001964) |
| 0.2 | 5.7 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.2 | 3.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.2 | 0.8 | GO:1990519 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.2 | 1.4 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.2 | 4.8 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.2 | 2.5 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 | 3.6 | GO:0060606 | neural tube closure(GO:0001843) tube closure(GO:0060606) |
| 0.2 | 1.3 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.2 | 0.7 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.2 | 0.5 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.2 | 3.3 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.2 | 0.5 | GO:1900060 | regulation of sphingolipid biosynthetic process(GO:0090153) negative regulation of sphingolipid biosynthetic process(GO:0090155) cellular sphingolipid homeostasis(GO:0090156) negative regulation of ceramide biosynthetic process(GO:1900060) regulation of membrane lipid metabolic process(GO:1905038) regulation of ceramide biosynthetic process(GO:2000303) |
| 0.2 | 2.2 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.2 | 4.8 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.2 | 3.5 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.2 | 4.3 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 9.5 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 16.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 3.9 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 2.3 | GO:0007286 | spermatid development(GO:0007286) |
| 0.1 | 0.4 | GO:2000058 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.1 | 6.7 | GO:0034620 | cellular response to unfolded protein(GO:0034620) |
| 0.1 | 3.8 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.1 | 2.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.6 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 1.5 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 3.0 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.1 | 3.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 1.5 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.1 | 1.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 2.9 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.1 | 4.3 | GO:0009755 | hormone-mediated signaling pathway(GO:0009755) |
| 0.1 | 1.2 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.8 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.1 | 4.8 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.1 | 2.9 | GO:0043405 | regulation of MAP kinase activity(GO:0043405) |
| 0.1 | 1.1 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 3.4 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 2.8 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 3.2 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 3.8 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.1 | 1.4 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.1 | 2.1 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 2.2 | GO:0010675 | regulation of cellular carbohydrate metabolic process(GO:0010675) regulation of glucose metabolic process(GO:0010906) |
| 0.1 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 1.3 | GO:0043409 | negative regulation of MAPK cascade(GO:0043409) |
| 0.1 | 2.7 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.1 | 1.0 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.1 | 1.6 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.1 | 5.2 | GO:0048738 | cardiac muscle tissue development(GO:0048738) |
| 0.1 | 1.3 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 1.0 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.0 | 4.1 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.9 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.3 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.0 | 1.9 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 2.3 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.0 | 7.5 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 1.4 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.6 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.9 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 3.3 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 1.3 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 1.0 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.9 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 2.0 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 2.2 | GO:0007268 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) |
| 0.0 | 4.1 | GO:0030097 | hemopoiesis(GO:0030097) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.2 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 2.0 | 5.9 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 1.0 | 6.9 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.9 | 5.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.8 | 9.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.8 | 3.2 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.6 | 11.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.6 | 8.2 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.5 | 2.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.5 | 4.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.5 | 5.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.4 | 2.6 | GO:0016234 | inclusion body(GO:0016234) |
| 0.4 | 8.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 2.7 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.3 | 3.0 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.3 | 1.8 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.2 | 11.6 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.2 | 1.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 5.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.2 | 1.3 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.2 | 2.8 | GO:0030315 | T-tubule(GO:0030315) |
| 0.2 | 0.5 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.2 | 3.2 | GO:0031672 | A band(GO:0031672) |
| 0.2 | 3.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.2 | 1.5 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.2 | 11.9 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.2 | 1.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 1.7 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 8.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 3.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 1.7 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 4.2 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.1 | 3.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 1.0 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 1.0 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 3.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 2.6 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 1.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 2.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 0.9 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 3.8 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 2.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 3.3 | GO:0030141 | secretory granule(GO:0030141) |
| 0.1 | 3.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 2.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 27.3 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.0 | 4.1 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.6 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 4.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 2.3 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 1.1 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 4.3 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.4 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 2.0 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.0 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 1.7 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 15.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.0 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 1.0 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.6 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.4 | GO:0035861 | site of double-strand break(GO:0035861) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.5 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
| 2.0 | 6.1 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 1.7 | 3.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 1.4 | 22.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 1.2 | 6.0 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 1.1 | 4.5 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 1.0 | 4.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 1.0 | 3.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.8 | 5.7 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.8 | 3.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.7 | 2.2 | GO:0008887 | glycerate kinase activity(GO:0008887) |
| 0.7 | 2.8 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.7 | 3.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.7 | 2.0 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.7 | 2.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.6 | 5.8 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.5 | 4.9 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.5 | 2.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.5 | 3.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.5 | 5.1 | GO:0001217 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.5 | 2.5 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.5 | 1.9 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.5 | 5.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.4 | 4.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.4 | 3.8 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.4 | 5.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.4 | 4.0 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.4 | 1.6 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.4 | 2.7 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.4 | 2.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.4 | 9.7 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.3 | 20.1 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.3 | 1.0 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.3 | 1.3 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.3 | 2.7 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.3 | 9.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.3 | 15.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.3 | 3.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 2.7 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.2 | 3.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 2.5 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.2 | 0.8 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.2 | 0.8 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.2 | 2.9 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 4.2 | GO:0004675 | transmembrane receptor protein serine/threonine kinase activity(GO:0004675) |
| 0.2 | 2.2 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.2 | 3.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 2.3 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.2 | 3.3 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.2 | 4.3 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 0.8 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 3.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.6 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 2.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 3.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 5.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 1.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.5 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 1.4 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 1.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.5 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.1 | 27.1 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 0.9 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 6.7 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.1 | 1.2 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 1.7 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.1 | 7.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 1.7 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.9 | GO:0004629 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 3.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 1.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 4.6 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 8.7 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 2.0 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.3 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 1.4 | GO:0015108 | chloride channel activity(GO:0005254) chloride transmembrane transporter activity(GO:0015108) |
| 0.0 | 3.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 2.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 2.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.4 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.0 | 1.0 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 1.9 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.0 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 1.3 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.0 | 0.6 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 16.2 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.3 | GO:0051787 | misfolded protein binding(GO:0051787) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.7 | 5.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.5 | 3.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.5 | 9.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.4 | 3.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.4 | 7.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.2 | 4.8 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.2 | 9.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.2 | 2.9 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 4.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 7.4 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 3.2 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.1 | 1.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 4.0 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 1.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.9 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 1.6 | 9.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.7 | 3.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.5 | 4.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.5 | 9.3 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.3 | 1.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.3 | 1.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.3 | 5.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.3 | 0.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.2 | 4.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 6.1 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.2 | 2.8 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.2 | 2.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 2.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 1.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.8 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 2.0 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 0.9 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.8 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.8 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 3.6 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.5 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |