Project
DANIO-CODE
Navigation
Downloads
Results for si:ch211-195d17.2
Z-value: 0.99
Transcription factors associated with si:ch211-195d17.2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
si_ch211-195d17.2
|
ENSDARG00000008119 | si_ch211-195d17.2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:ch211-195d17.2 | dr10_dc_chr20_-_29780783_29780926 | -0.37 | 1.6e-01 | Click! |
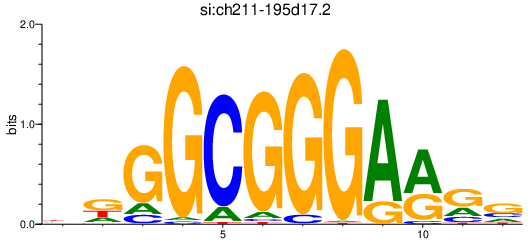
Activity profile of si:ch211-195d17.2 motif
Sorted Z-values of si:ch211-195d17.2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of si:ch211-195d17.2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_21485280 | 1.62 |
ENSDART00000066896
|
syt1a
|
synaptotagmin Ia |
| chr8_-_46963909 | 1.49 |
|
|
|
| chr25_+_34627031 | 1.47 |
ENSDART00000154377
|
HIST1H2BA (1 of many)
|
histone cluster 1 H2B family member a |
| chr25_-_34619603 | 1.20 |
ENSDART00000114767
|
FQ312024.1
|
ENSDARG00000076129 |
| chr7_-_6309265 | 1.14 |
ENSDART00000172825
|
FP325123.5
|
Histone H3.2 |
| chr25_-_35820568 | 1.09 |
ENSDART00000152766
|
CR354435.4
|
Histone H2B 1/2 |
| chr25_+_34562216 | 0.99 |
ENSDART00000154655
|
CU302436.4
|
ENSDARG00000092743 |
| chr7_-_6226607 | 0.98 |
ENSDART00000129239
|
HIST2H2AB (1 of many)
|
histone cluster 2 H2A family member b |
| chr25_+_34553079 | 0.98 |
ENSDART00000156838
|
zgc:112234
|
zgc:112234 |
| chr25_-_35817806 | 0.97 |
ENSDART00000129969
|
ENSDARG00000086604
|
ENSDARG00000086604 |
| chr7_+_5839103 | 0.96 |
ENSDART00000145370
|
zgc:112234
|
zgc:112234 |
| chr7_+_73601671 | 0.89 |
ENSDART00000163075
|
zgc:173552
|
zgc:173552 |
| chr7_-_73631286 | 0.83 |
ENSDART00000129254
|
zgc:173552
|
zgc:173552 |
| chr7_-_6209659 | 0.79 |
ENSDART00000173032
|
CU457819.4
|
Histone H3.2 |
| chr7_+_5855024 | 0.75 |
ENSDART00000173105
|
si:dkey-23a13.21
|
si:dkey-23a13.21 |
| chr25_-_34542816 | 0.70 |
ENSDART00000155060
|
zgc:110434
|
zgc:110434 |
| KN150123v1_-_37214 | 0.67 |
|
|
|
| chr21_+_26660833 | 0.67 |
ENSDART00000004109
|
gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr3_-_32202533 | 0.66 |
ENSDART00000155757
|
si:dkey-16p21.8
|
si:dkey-16p21.8 |
| chr19_-_9553977 | 0.64 |
ENSDART00000045565
|
vamp1
|
vesicle-associated membrane protein 1 |
| chr11_-_41357639 | 0.63 |
ENSDART00000055709
|
her2
|
hairy-related 2 |
| chr7_+_73594790 | 0.63 |
ENSDART00000123081
|
zgc:173552
|
zgc:173552 |
| chr20_+_27194162 | 0.63 |
ENSDART00000024595
|
ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 |
| chr19_+_48731885 | 0.62 |
|
|
|
| chr17_-_117745 | 0.61 |
|
|
|
| chr7_+_5854890 | 0.61 |
ENSDART00000173105
|
si:dkey-23a13.21
|
si:dkey-23a13.21 |
| chr14_+_22889826 | 0.60 |
ENSDART00000170356
|
enox2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr7_-_29452646 | 0.59 |
ENSDART00000112651
|
vps13c
|
vacuolar protein sorting 13 homolog C (S. cerevisiae) |
| chr3_-_19050721 | 0.59 |
ENSDART00000131503
|
rfx1a
|
regulatory factor X, 1a (influences HLA class II expression) |
| chr19_+_24398194 | 0.58 |
ENSDART00000080673
|
syt11a
|
synaptotagmin XIa |
| chr16_+_17481157 | 0.56 |
ENSDART00000173448
|
fam131bb
|
family with sequence similarity 131, member Bb |
| chr4_+_21485372 | 0.55 |
ENSDART00000041861
|
syt1a
|
synaptotagmin Ia |
| chr11_+_40567461 | 0.55 |
ENSDART00000160023
|
errfi1a
|
ERBB receptor feedback inhibitor 1a |
| KN150648v1_+_877 | 0.55 |
|
|
|
| chr7_-_68417190 | 0.55 |
|
|
|
| chr5_-_63031745 | 0.54 |
|
|
|
| chr12_+_47450923 | 0.54 |
ENSDART00000105328
|
fmn2b
|
formin 2b |
| chr13_+_31271568 | 0.54 |
ENSDART00000019202
|
tdrd9
|
tudor domain containing 9 |
| KN149861v1_-_5543 | 0.53 |
|
|
|
| chr24_-_2918801 | 0.52 |
ENSDART00000164776
|
fam69c
|
family with sequence similarity 69, member C |
| chr7_-_26035308 | 0.51 |
ENSDART00000131906
|
zgc:77439
|
zgc:77439 |
| chr11_+_24687813 | 0.49 |
ENSDART00000131431
|
sulf2a
|
sulfatase 2a |
| chr23_+_4955111 | 0.48 |
ENSDART00000023537
|
tnnc1a
|
troponin C type 1a (slow) |
| chr19_-_48731766 | 0.48 |
ENSDART00000170726
|
si:ch73-359m17.2
|
si:ch73-359m17.2 |
| chr25_-_34605715 | 0.48 |
ENSDART00000099859
|
CU302436.2
|
ENSDARG00000068928 |
| chr24_+_10273081 | 0.47 |
ENSDART00000111014
|
myca
|
MYC proto-oncogene, bHLH transcription factor a |
| chr17_-_117828 | 0.47 |
|
|
|
| chr16_-_13063634 | 0.47 |
ENSDART00000103894
|
cacng8b
|
calcium channel, voltage-dependent, gamma subunit 8b |
| chr7_+_39487676 | 0.46 |
ENSDART00000111278
|
sorcs2
|
sortilin-related VPS10 domain containing receptor 2 |
| chr18_-_45750 | 0.46 |
ENSDART00000148821
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr7_+_25587096 | 0.45 |
ENSDART00000174782
|
mtmr1a
|
myotubularin related protein 1a |
| chr17_+_2551397 | 0.45 |
ENSDART00000178759
|
kcnk10b
|
potassium channel, subfamily K, member 10b |
| chr1_+_44113013 | 0.44 |
ENSDART00000059227
|
ndufs8a
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8a |
| chr7_-_34854886 | 0.44 |
ENSDART00000141211
|
hsd11b2
|
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr6_-_10728582 | 0.44 |
ENSDART00000151102
|
notum2
|
notum pectinacetylesterase 2 |
| chr4_+_248611 | 0.43 |
|
|
|
| chr25_-_6501787 | 0.42 |
ENSDART00000112782
|
cspg4
|
chondroitin sulfate proteoglycan 4 |
| chr2_-_44330405 | 0.42 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr20_-_54160322 | 0.42 |
ENSDART00000153435
|
ppp2r5cb
|
protein phosphatase 2, regulatory subunit B', gamma b |
| chr19_-_44495755 | 0.41 |
ENSDART00000132800
|
rad21b
|
RAD21 cohesin complex component b |
| chr7_-_29300402 | 0.41 |
ENSDART00000099477
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr17_+_22359660 | 0.41 |
ENSDART00000162670
|
slc8a1b
|
solute carrier family 8 (sodium/calcium exchanger), member 1b |
| chr25_+_7000082 | 0.41 |
|
|
|
| chr6_-_32718634 | 0.40 |
ENSDART00000175666
|
mafaa
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog Aa |
| chr22_-_21021866 | 0.38 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr12_+_25132396 | 0.38 |
ENSDART00000139362
|
mta3
|
metastasis associated 1 family, member 3 |
| chr19_-_32900108 | 0.38 |
ENSDART00000050130
|
gmpr
|
guanosine monophosphate reductase |
| chr7_+_25587183 | 0.38 |
ENSDART00000148780
|
mtmr1a
|
myotubularin related protein 1a |
| chr10_+_20151082 | 0.37 |
ENSDART00000142708
|
dmtn
|
dematin actin binding protein |
| chr16_+_24927745 | 0.37 |
ENSDART00000157333
|
si:dkey-79d12.6
|
si:dkey-79d12.6 |
| chr9_+_8990576 | 0.37 |
ENSDART00000133899
|
ube2al
|
ubiquitin conjugating enzyme E2 A, like |
| chr18_+_49230636 | 0.37 |
ENSDART00000167609
ENSDART00000135026 ENSDART00000171618 |
si:ch211-136a13.1
|
si:ch211-136a13.1 |
| chr5_-_66686945 | 0.37 |
ENSDART00000133438
|
unga
|
uracil DNA glycosylase a |
| chr11_-_27378184 | 0.36 |
ENSDART00000157337
|
CR931782.1
|
ENSDARG00000097455 |
| chr2_-_4277605 | 0.36 |
|
|
|
| chr11_-_1264715 | 0.36 |
ENSDART00000167818
|
atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr5_+_29578638 | 0.36 |
ENSDART00000134624
|
adamts15a
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15a |
| KN150640v1_+_5728 | 0.36 |
|
|
|
| chr11_-_6960107 | 0.36 |
ENSDART00000171255
|
COMP
|
cartilage oligomeric matrix protein |
| chr13_+_28997676 | 0.36 |
ENSDART00000109546
|
unc5b
|
unc-5 netrin receptor B |
| chr20_-_32543497 | 0.35 |
ENSDART00000026635
|
nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr21_-_34623741 | 0.35 |
ENSDART00000023038
|
dacha
|
dachshund a |
| chr14_-_24304373 | 0.35 |
|
|
|
| chr3_+_21928483 | 0.35 |
ENSDART00000155739
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr4_-_25080470 | 0.35 |
ENSDART00000179640
|
gata3
|
GATA binding protein 3 |
| chr23_+_36207653 | 0.35 |
ENSDART00000053267
|
hnrnpa1b
|
heterogeneous nuclear ribonucleoprotein A1b |
| chr9_+_17779858 | 0.34 |
ENSDART00000013111
|
dgkh
|
diacylglycerol kinase, eta |
| chr17_-_52735250 | 0.34 |
|
|
|
| chr3_+_25914076 | 0.34 |
ENSDART00000133523
|
hmgxb4a
|
HMG box domain containing 4a |
| chr12_+_25132724 | 0.33 |
ENSDART00000127454
ENSDART00000122665 |
mta3
|
metastasis associated 1 family, member 3 |
| chr20_+_15119754 | 0.33 |
ENSDART00000039345
|
myoc
|
myocilin |
| chr1_-_22466834 | 0.33 |
ENSDART00000144208
|
adgrl3.1
|
adhesion G protein-coupled receptor L3.1 |
| chr16_-_12896908 | 0.33 |
ENSDART00000139916
|
foxj2
|
forkhead box J2 |
| chr1_+_26418707 | 0.33 |
ENSDART00000161169
|
bnc2
|
basonuclin 2 |
| chr12_+_47451037 | 0.33 |
ENSDART00000105328
|
fmn2b
|
formin 2b |
| chr9_-_21257082 | 0.33 |
ENSDART00000124533
|
tbx15
|
T-box 15 |
| chr16_-_9108885 | 0.33 |
ENSDART00000153785
|
triob
|
trio Rho guanine nucleotide exchange factor b |
| chr18_+_18115168 | 0.33 |
ENSDART00000145342
|
cbln1
|
cerebellin 1 precursor |
| chr7_-_6215188 | 0.32 |
ENSDART00000159542
|
zgc:112234
|
zgc:112234 |
| chr23_+_31888903 | 0.32 |
ENSDART00000075730
|
myb
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr7_+_73408688 | 0.32 |
ENSDART00000159745
|
PCP4L1
|
Purkinje cell protein 4 like 1 |
| chr2_-_2109125 | 0.31 |
ENSDART00000101033
|
pth1ra
|
parathyroid hormone 1 receptor a |
| chr14_+_11457009 | 0.31 |
ENSDART00000110004
|
frmpd3
|
FERM and PDZ domain containing 3 |
| chr11_-_13069266 | 0.31 |
ENSDART00000169052
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr21_+_44203036 | 0.31 |
|
|
|
| chr23_-_18131105 | 0.31 |
ENSDART00000173102
|
zgc:92287
|
zgc:92287 |
| chr8_+_39400230 | 0.30 |
|
|
|
| chr17_+_13654337 | 0.30 |
ENSDART00000171689
|
lrfn5a
|
leucine rich repeat and fibronectin type III domain containing 5a |
| chr17_+_51272621 | 0.30 |
|
|
|
| chr3_-_27944852 | 0.30 |
ENSDART00000122037
|
rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1 |
| chr21_+_23916512 | 0.30 |
ENSDART00000145541
ENSDART00000065599 ENSDART00000112869 |
cadm1a
|
cell adhesion molecule 1a |
| chr7_-_73620490 | 0.29 |
ENSDART00000158891
ENSDART00000111622 |
FP236812.3
|
ENSDARG00000079652 |
| chr5_-_21384509 | 0.29 |
ENSDART00000020725
|
smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr1_+_40428827 | 0.29 |
ENSDART00000145272
|
lrpap1
|
low density lipoprotein receptor-related protein associated protein 1 |
| chr5_-_70991958 | 0.29 |
ENSDART00000011955
ENSDART00000160380 |
gpsm1b
|
G protein signaling modulator 1b |
| chr2_+_32033028 | 0.29 |
ENSDART00000005143
|
mycb
|
MYC proto-oncogene, bHLH transcription factor b |
| chr5_-_40707316 | 0.29 |
ENSDART00000161932
|
npr3
|
natriuretic peptide receptor 3 |
| chr1_-_19580295 | 0.29 |
ENSDART00000146084
|
ndst3
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
| chr14_+_43599635 | 0.29 |
ENSDART00000155539
|
CR786577.1
|
ENSDARG00000097875 |
| chr24_-_13204960 | 0.29 |
ENSDART00000134482
|
terf1
|
telomeric repeat binding factor (NIMA-interacting) 1 |
| chr12_+_18914438 | 0.28 |
ENSDART00000153086
|
kctd17
|
potassium channel tetramerization domain containing 17 |
| chr11_+_41801746 | 0.28 |
ENSDART00000173252
|
camta1
|
calmodulin binding transcription activator 1 |
| chr3_-_10667865 | 0.27 |
ENSDART00000130761
ENSDART00000156617 |
map2k4a
|
mitogen-activated protein kinase kinase 4a |
| chr25_-_34640899 | 0.27 |
ENSDART00000157137
|
ENSDARG00000070254
|
ENSDARG00000070254 |
| chr15_+_21399299 | 0.27 |
ENSDART00000154914
|
BX663520.1
|
ENSDARG00000097950 |
| chr3_-_45369476 | 0.27 |
ENSDART00000167179
|
asf1ba
|
anti-silencing function 1Ba histone chaperone |
| chr8_+_29626848 | 0.27 |
ENSDART00000139029
ENSDART00000091409 |
smarcad1a
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 a |
| chr7_-_29300448 | 0.27 |
ENSDART00000019140
|
rorab
|
RAR-related orphan receptor A, paralog b |
| chr25_+_34557335 | 0.26 |
ENSDART00000089844
|
zgc:113983
|
zgc:113983 |
| chr20_-_54364559 | 0.26 |
|
|
|
| chr3_-_15918262 | 0.26 |
ENSDART00000157315
|
ENSDARG00000097322
|
ENSDARG00000097322 |
| chr25_+_32114076 | 0.26 |
ENSDART00000156145
|
scaper
|
S-phase cyclin A-associated protein in the ER |
| chr11_-_23226859 | 0.26 |
ENSDART00000159221
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr23_-_18131209 | 0.26 |
ENSDART00000173102
|
zgc:92287
|
zgc:92287 |
| chr23_+_3788435 | 0.26 |
ENSDART00000141782
|
smim29
|
small integral membrane protein 29 |
| chr5_-_66687280 | 0.26 |
ENSDART00000062359
|
unga
|
uracil DNA glycosylase a |
| chr12_+_16235258 | 0.26 |
ENSDART00000141169
|
ppp1r3cb
|
protein phosphatase 1, regulatory subunit 3Cb |
| chr5_-_66687387 | 0.26 |
ENSDART00000062359
|
unga
|
uracil DNA glycosylase a |
| chr2_-_4277516 | 0.26 |
|
|
|
| chr23_+_20592121 | 0.26 |
ENSDART00000114246
|
adnpb
|
activity-dependent neuroprotector homeobox b |
| chr18_+_46164175 | 0.26 |
ENSDART00000113545
ENSDART00000062049 |
zgc:113340
|
zgc:113340 |
| chr8_+_28046911 | 0.26 |
ENSDART00000078533
|
kcnd3
|
potassium voltage-gated channel, Shal-related subfamily, member 3 |
| chr10_-_40905193 | 0.26 |
ENSDART00000172089
|
pcna
|
proliferating cell nuclear antigen |
| chr21_+_43674754 | 0.25 |
ENSDART00000136025
|
tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr22_-_22276825 | 0.25 |
ENSDART00000111711
|
chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr4_+_22851260 | 0.25 |
ENSDART00000174705
|
sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr4_-_66538843 | 0.25 |
ENSDART00000165173
|
BX548011.1
|
ENSDARG00000103357 |
| chr7_+_5791368 | 0.25 |
ENSDART00000160575
|
HIST1H3I
|
Histone H3.2 |
| chr25_-_34605630 | 0.25 |
ENSDART00000099859
|
CU302436.2
|
ENSDARG00000068928 |
| chr3_+_21928413 | 0.24 |
ENSDART00000122782
|
kansl1b
|
KAT8 regulatory NSL complex subunit 1b |
| chr7_+_73408746 | 0.24 |
ENSDART00000159745
|
PCP4L1
|
Purkinje cell protein 4 like 1 |
| chr10_-_18510699 | 0.24 |
|
|
|
| chr7_+_24610446 | 0.24 |
ENSDART00000058843
|
krcp
|
kelch repeat-containing protein |
| chr4_-_1809690 | 0.24 |
|
|
|
| chr3_-_43730944 | 0.24 |
|
|
|
| chr17_+_14776302 | 0.23 |
ENSDART00000154229
|
RTRAF
|
zgc:56576 |
| KN149707v1_+_4014 | 0.23 |
ENSDART00000168516
|
cdo1
|
cysteine dioxygenase, type I |
| chr3_+_16115708 | 0.23 |
ENSDART00000122519
|
st8sia6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
| chr14_-_24304628 | 0.23 |
|
|
|
| chr15_+_21399127 | 0.23 |
ENSDART00000154914
|
BX663520.1
|
ENSDARG00000097950 |
| chr2_-_55971357 | 0.23 |
ENSDART00000154701
ENSDART00000154107 |
si:ch211-178n15.1
|
si:ch211-178n15.1 |
| chr1_-_30298698 | 0.23 |
|
|
|
| chr22_+_3994527 | 0.23 |
ENSDART00000166768
|
timm44
|
translocase of inner mitochondrial membrane 44 homolog (yeast) |
| chr18_+_9213713 | 0.23 |
ENSDART00000127469
ENSDART00000101192 |
sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr7_+_60054116 | 0.22 |
ENSDART00000145201
|
ppp1r14bb
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Bb |
| chr7_+_44106327 | 0.22 |
ENSDART00000108766
ENSDART00000111441 |
cdh5
|
cadherin 5 |
| chr8_+_5222065 | 0.22 |
ENSDART00000035676
|
bnip3la
|
BCL2/adenovirus E1B interacting protein 3-like a |
| chr8_+_54171992 | 0.22 |
ENSDART00000122692
|
CABZ01112317.1
|
ENSDARG00000086057 |
| chr18_-_16134320 | 0.22 |
ENSDART00000061189
|
sspn
|
sarcospan (Kras oncogene-associated gene) |
| chr19_-_43037791 | 0.22 |
ENSDART00000004392
|
fkbp9
|
FK506 binding protein 9 |
| chr6_-_30818840 | 0.22 |
|
|
|
| chr11_-_6209253 | 0.22 |
ENSDART00000150199
|
pole4
|
polymerase (DNA-directed), epsilon 4, accessory subunit |
| chr13_+_11741597 | 0.22 |
|
|
|
| chr8_-_32497168 | 0.22 |
ENSDART00000164826
|
RFESD (1 of many)
|
Rieske Fe-S domain containing |
| chr8_+_29626894 | 0.22 |
ENSDART00000139029
ENSDART00000091409 |
smarcad1a
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 a |
| chr1_+_40428722 | 0.22 |
ENSDART00000145272
|
lrpap1
|
low density lipoprotein receptor-related protein associated protein 1 |
| chr25_+_17327279 | 0.22 |
ENSDART00000061738
|
elmo3
|
engulfment and cell motility 3 |
| chr16_-_22397159 | 0.22 |
|
|
|
| chr25_-_35817715 | 0.21 |
ENSDART00000129969
|
ENSDARG00000086604
|
ENSDARG00000086604 |
| chr20_-_47521258 | 0.21 |
|
|
|
| chr3_+_53913303 | 0.21 |
ENSDART00000109894
|
olfm2a
|
olfactomedin 2a |
| chr18_-_16134258 | 0.21 |
ENSDART00000061189
|
sspn
|
sarcospan (Kras oncogene-associated gene) |
| chr19_+_31046291 | 0.21 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr5_-_37784878 | 0.21 |
ENSDART00000051233
|
mink1
|
misshapen-like kinase 1 |
| chr25_-_34622553 | 0.20 |
ENSDART00000125128
|
FP236630.1
|
ENSDARG00000086304 |
| chr7_+_71096788 | 0.20 |
ENSDART00000161871
|
sod3a
|
superoxide dismutase 3, extracellular a |
| chr19_-_31378033 | 0.20 |
|
|
|
| chr13_-_33527029 | 0.20 |
|
|
|
| chr5_-_29935062 | 0.20 |
ENSDART00000040328
|
h2afx
|
H2A histone family, member X |
| chr24_+_37802000 | 0.20 |
|
|
|
| chr3_-_43730993 | 0.20 |
|
|
|
| chr11_+_2786083 | 0.20 |
ENSDART00000172837
|
kif21b
|
kinesin family member 21B |
| chr6_+_49927433 | 0.20 |
ENSDART00000018523
|
ahcy
|
adenosylhomocysteinase |
| chr22_+_21300705 | 0.19 |
|
|
|
| chr11_-_21143897 | 0.19 |
ENSDART00000163008
|
RASSF5
|
Ras association domain family member 5 |
| chr7_-_19989384 | 0.19 |
ENSDART00000173619
|
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr3_+_59608335 | 0.19 |
ENSDART00000064311
|
arhgdia
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr10_-_35598016 | 0.19 |
ENSDART00000162139
|
si:ch211-244c8.4
|
si:ch211-244c8.4 |
| chr7_-_26035541 | 0.18 |
ENSDART00000057288
|
zgc:77439
|
zgc:77439 |
| chr10_-_22834248 | 0.18 |
ENSDART00000079469
|
pcolcea
|
procollagen C-endopeptidase enhancer a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 0.8 | GO:0097510 | base-excision repair, AP site formation via deaminated base removal(GO:0097510) |
| 0.2 | 0.6 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.2 | 0.5 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.1 | 0.6 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.1 | 0.5 | GO:0010915 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) regulation of lipoprotein particle clearance(GO:0010984) negative regulation of lipoprotein particle clearance(GO:0010985) |
| 0.1 | 0.6 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.1 | 0.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.1 | 0.6 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.1 | 0.3 | GO:0030852 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.1 | 0.3 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 0.1 | 0.3 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.1 | 0.3 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 0.5 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.4 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.1 | 0.2 | GO:0002164 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.1 | 0.2 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.1 | 0.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.1 | 0.4 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 0.1 | 0.6 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 0.6 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.5 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.4 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.7 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.3 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.7 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.5 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.0 | 0.2 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.0 | 0.2 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.2 | GO:1990748 | response to superoxide(GO:0000303) response to oxygen radical(GO:0000305) removal of superoxide radicals(GO:0019430) cellular response to oxygen radical(GO:0071450) cellular response to superoxide(GO:0071451) cellular oxidant detoxification(GO:0098869) cellular detoxification(GO:1990748) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.2 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.0 | 0.4 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.2 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.3 | GO:0014059 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) |
| 0.0 | 0.4 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.8 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.3 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.0 | 0.1 | GO:0099639 | neurotransmitter receptor transport, endosome to postsynaptic membrane(GO:0098887) endosome to plasma membrane protein transport(GO:0099638) neurotransmitter receptor transport, endosome to plasma membrane(GO:0099639) |
| 0.0 | 0.3 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.0 | 0.5 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.4 | GO:0097581 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 1.0 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.3 | GO:0060038 | cardiac muscle cell proliferation(GO:0060038) |
| 0.0 | 0.3 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.3 | GO:0039020 | pronephric nephron tubule development(GO:0039020) |
| 0.0 | 0.1 | GO:0010867 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.3 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 0.1 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.7 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.2 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 0.4 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.1 | 0.7 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.6 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.5 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 0.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.5 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 2.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.1 | GO:0044326 | dendritic spine neck(GO:0044326) dendritic filopodium(GO:1902737) |
| 0.0 | 0.5 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.3 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.6 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.4 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.2 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.2 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 0.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.7 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 0.4 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 2.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.3 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.1 | 0.8 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 0.3 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.1 | 0.6 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 0.7 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.1 | 0.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.5 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.4 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.3 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.2 | GO:0016801 | hydrolase activity, acting on ether bonds(GO:0016801) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.4 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.4 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.0 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 0.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 2.8 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.9 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.3 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.2 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 0.9 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 0.2 | GO:0008198 | ferrous iron binding(GO:0008198) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.3 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.5 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.2 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.6 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 0.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |