Project
DANIO-CODE
Navigation
Downloads
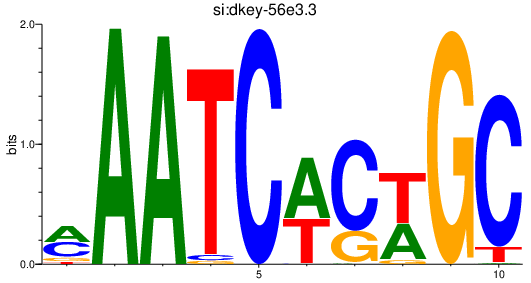
Results for si:dkey-56e3.3
Z-value: 2.52
Transcription factors associated with si:dkey-56e3.3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
si_dkey-56e3.3
|
ENSDARG00000075194 | si_dkey-56e3.3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| si:dkey-56e3.3 | dr10_dc_chr1_+_40894908_40895077 | -0.87 | 1.1e-05 | Click! |
Activity profile of si:dkey-56e3.3 motif
Sorted Z-values of si:dkey-56e3.3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of si:dkey-56e3.3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr23_-_9833729 | 6.89 |
ENSDART00000045126
|
lama5
|
laminin, alpha 5 |
| chr22_-_37414940 | 6.71 |
ENSDART00000104493
|
sox2
|
SRY (sex determining region Y)-box 2 |
| chr17_+_15425559 | 6.65 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr6_-_47842137 | 6.12 |
ENSDART00000141986
|
lrig2
|
leucine-rich repeats and immunoglobulin-like domains 2 |
| chr23_-_35595271 | 6.09 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr3_-_48865474 | 5.97 |
ENSDART00000133036
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr2_-_47578695 | 5.79 |
ENSDART00000014350
ENSDART00000038828 |
pax3a
|
paired box 3a |
| chr4_-_9721600 | 5.76 |
ENSDART00000067190
|
tspan9b
|
tetraspanin 9b |
| chr19_-_28546417 | 5.69 |
ENSDART00000130922
ENSDART00000079114 |
irx1b
|
iroquois homeobox 1b |
| chr14_-_17258072 | 5.50 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor-like 1a |
| chr8_-_23172938 | 5.42 |
ENSDART00000141090
|
si:ch211-196c10.15
|
si:ch211-196c10.15 |
| chr22_+_15995914 | 5.10 |
ENSDART00000062618
|
serpinc1
|
serpin peptidase inhibitor, clade C (antithrombin), member 1 |
| chr16_+_17705704 | 5.08 |
|
|
|
| chr12_+_31558667 | 5.05 |
ENSDART00000152971
|
dnmbp
|
dynamin binding protein |
| chr19_+_31817286 | 5.05 |
ENSDART00000078459
|
tmem55a
|
transmembrane protein 55A |
| chr2_+_27344633 | 5.04 |
ENSDART00000178275
|
cdh7
|
cadherin 7, type 2 |
| KN149998v1_+_54953 | 5.02 |
|
|
|
| chr2_-_24898678 | 4.94 |
ENSDART00000145145
|
cnn2
|
calponin 2 |
| chr8_+_39573104 | 4.90 |
ENSDART00000158498
ENSDART00000041634 ENSDART00000169542 |
msi1
|
musashi RNA-binding protein 1 |
| chr7_-_70219217 | 4.90 |
ENSDART00000097710
|
ppargc1a
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
| chr1_+_16681778 | 4.86 |
|
|
|
| chr18_-_14891913 | 4.81 |
ENSDART00000018502
|
mapk12a
|
mitogen-activated protein kinase 12a |
| chr16_+_737615 | 4.73 |
ENSDART00000161774
|
irx1a
|
iroquois homeobox 1a |
| chr6_+_35378839 | 4.70 |
ENSDART00000102483
ENSDART00000133783 |
rgs4
|
regulator of G protein signaling 4 |
| chr2_+_33057931 | 4.66 |
|
|
|
| chr1_-_46398498 | 4.51 |
ENSDART00000000087
|
itsn1
|
intersectin 1 (SH3 domain protein) |
| chr7_+_25649559 | 4.48 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr4_-_5325164 | 4.23 |
ENSDART00000067375
|
osgep
|
O-sialoglycoprotein endopeptidase |
| chr3_-_32727588 | 4.22 |
ENSDART00000158916
|
si:dkey-16l2.20
|
si:dkey-16l2.20 |
| chr9_+_48309684 | 4.21 |
ENSDART00000177287
ENSDART00000113883 |
ENSDARG00000041339
|
ENSDARG00000041339 |
| chr6_-_23455346 | 4.20 |
ENSDART00000157527
ENSDART00000168882 ENSDART00000171384 |
ntn1a
|
netrin 1a |
| chr7_-_12715948 | 4.19 |
ENSDART00000173115
|
rplp2l
|
ribosomal protein, large P2, like |
| chr25_-_7346766 | 4.02 |
ENSDART00000170050
|
cdkn1cb
|
cyclin-dependent kinase inhibitor 1Cb |
| chr25_-_33920256 | 4.00 |
|
|
|
| chr8_-_19364689 | 3.95 |
ENSDART00000014183
|
colgalt2
|
collagen beta(1-O)galactosyltransferase 2 |
| chr8_+_29953399 | 3.87 |
ENSDART00000149372
ENSDART00000007640 |
ptch1
ptch1
|
patched 1 patched 1 |
| chr16_+_21109486 | 3.87 |
ENSDART00000079383
|
hoxa9b
|
homeobox A9b |
| chr21_-_37286942 | 3.85 |
ENSDART00000100310
|
dbn1
|
drebrin 1 |
| chr23_-_41756325 | 3.77 |
ENSDART00000146808
|
si:ch73-184c24.1
|
si:ch73-184c24.1 |
| chr14_+_33382973 | 3.74 |
ENSDART00000132488
|
apln
|
apelin |
| chr13_+_13550656 | 3.69 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr6_-_15526547 | 3.69 |
ENSDART00000038133
|
trim63a
|
tripartite motif containing 63a |
| chr17_+_37268262 | 3.68 |
ENSDART00000104009
|
slc30a1b
|
solute carrier family 30 (zinc transporter), member 1b |
| chr8_+_15987710 | 3.66 |
ENSDART00000165141
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr11_-_25962291 | 3.62 |
|
|
|
| chr20_-_19646761 | 3.60 |
|
|
|
| chr21_-_41286846 | 3.56 |
ENSDART00000167339
|
msx2b
|
muscle segment homeobox 2b |
| chr15_+_28435937 | 3.54 |
ENSDART00000142298
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr8_+_53118919 | 3.54 |
|
|
|
| chr22_-_5624894 | 3.51 |
ENSDART00000165221
|
mcm2
|
minichromosome maintenance complex component 2 |
| chr16_+_50189177 | 3.50 |
ENSDART00000163565
|
plcl2
|
phospholipase C-like 2 |
| chr23_+_23305483 | 3.49 |
ENSDART00000126479
|
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr19_+_16318256 | 3.49 |
ENSDART00000137189
|
ptprua
|
protein tyrosine phosphatase, receptor type, U, a |
| KN150617v1_+_66361 | 3.48 |
ENSDART00000159503
|
fzd4
|
frizzled class receptor 4 |
| chr23_+_36832325 | 3.48 |
|
|
|
| chr24_-_6048914 | 3.43 |
ENSDART00000146830
ENSDART00000021981 |
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr23_-_10202347 | 3.41 |
ENSDART00000142442
|
plxnb1a
|
plexin b1a |
| chr2_+_4233224 | 3.41 |
ENSDART00000163737
|
mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr15_-_28547610 | 3.35 |
ENSDART00000057696
|
git1
|
G protein-coupled receptor kinase interacting ArfGAP 1 |
| chr4_-_76594677 | 3.34 |
ENSDART00000049170
|
zgc:85975
|
zgc:85975 |
| chr6_+_19795100 | 3.33 |
|
|
|
| chr17_+_12544451 | 3.30 |
ENSDART00000064509
ENSDART00000136830 |
stmn4l
|
stathmin-like 4, like |
| chr24_+_4946542 | 3.30 |
ENSDART00000045813
|
zic4
|
zic family member 4 |
| chr21_+_38264846 | 3.25 |
ENSDART00000065159
|
zgc:158291
|
zgc:158291 |
| chr8_-_22945616 | 3.24 |
|
|
|
| chr18_+_19659195 | 3.23 |
ENSDART00000100569
|
smad6b
|
SMAD family member 6b |
| chr16_+_19730756 | 3.21 |
ENSDART00000112894
|
sp8b
|
sp8 transcription factor b |
| chr2_-_42279180 | 3.20 |
ENSDART00000047055
|
trim55a
|
tripartite motif containing 55a |
| chr24_+_10276926 | 3.20 |
ENSDART00000145771
ENSDART00000157350 |
CT573344.1
|
ENSDARG00000093108 |
| chr12_-_30988279 | 3.15 |
ENSDART00000122972
ENSDART00000005562 ENSDART00000031408 ENSDART00000125046 ENSDART00000009237 |
tcf7l2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr7_+_57423271 | 3.15 |
ENSDART00000056466
|
camk2d1
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 1 |
| chr13_+_30743690 | 3.11 |
ENSDART00000134809
|
ercc6
|
excision repair cross-complementation group 6 |
| chr23_+_24711233 | 3.10 |
|
|
|
| chr15_-_19789174 | 3.10 |
ENSDART00000152345
ENSDART00000047643 ENSDART00000163435 |
sytl2b
|
synaptotagmin-like 2b |
| chr18_-_49121584 | 3.07 |
ENSDART00000174157
|
ENSDARG00000099137
|
ENSDARG00000099137 |
| chr9_-_1949904 | 3.03 |
ENSDART00000082355
|
hoxd4a
|
homeobox D4a |
| chr21_+_10775209 | 3.03 |
|
|
|
| chr25_+_36872560 | 3.03 |
ENSDART00000163178
|
slc10a3
|
solute carrier family 10, member 3 |
| chr23_+_35996491 | 3.00 |
ENSDART00000127384
|
hoxc9a
|
homeobox C9a |
| chr3_-_21217799 | 2.96 |
ENSDART00000114906
|
fam171a2a
|
family with sequence similarity 171, member A2a |
| KN149726v1_+_1094 | 2.94 |
|
|
|
| chr2_-_32468715 | 2.93 |
|
|
|
| chr15_-_14616083 | 2.92 |
ENSDART00000171169
|
numbl
|
numb homolog (Drosophila)-like |
| chr12_+_25509394 | 2.90 |
ENSDART00000077157
|
six3b
|
SIX homeobox 3b |
| chr1_+_30992553 | 2.89 |
ENSDART00000112333
|
cnnm2b
|
cyclin and CBS domain divalent metal cation transport mediator 2b |
| chr17_-_26893412 | 2.88 |
ENSDART00000045842
|
rcan3
|
regulator of calcineurin 3 |
| chr4_-_19705018 | 2.88 |
ENSDART00000100974
|
snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr23_+_22729939 | 2.84 |
ENSDART00000009337
|
eno1a
|
enolase 1a, (alpha) |
| chr7_+_24770873 | 2.80 |
ENSDART00000165314
|
AL954715.1
|
ENSDARG00000103682 |
| chr12_+_34852049 | 2.80 |
ENSDART00000027034
|
qki2
|
QKI, KH domain containing, RNA binding 2 |
| chr10_+_33926312 | 2.77 |
ENSDART00000174730
|
BX569785.2
|
ENSDARG00000106629 |
| chr13_+_21649283 | 2.77 |
ENSDART00000021556
|
si:ch211-51a6.2
|
si:ch211-51a6.2 |
| chr3_-_5318289 | 2.76 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr15_-_10345337 | 2.75 |
|
|
|
| chr23_-_28367816 | 2.75 |
ENSDART00000003548
|
znf385a
|
zinc finger protein 385A |
| chr16_+_19730958 | 2.71 |
ENSDART00000139357
|
sp8b
|
sp8 transcription factor b |
| chr16_+_19730820 | 2.71 |
ENSDART00000079201
|
sp8b
|
sp8 transcription factor b |
| chr24_-_40968409 | 2.70 |
ENSDART00000169315
ENSDART00000171543 |
smyhc1
|
slow myosin heavy chain 1 |
| chr3_+_49277125 | 2.67 |
ENSDART00000161724
|
gas7a
|
growth arrest-specific 7a |
| chr4_+_22759177 | 2.66 |
ENSDART00000146272
|
ndufb2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2 |
| chr8_+_33010788 | 2.65 |
ENSDART00000172672
|
angptl2b
|
angiopoietin-like 2b |
| chr18_+_23891068 | 2.65 |
ENSDART00000146490
|
BX569787.1
|
ENSDARG00000092402 |
| chr25_-_13737344 | 2.64 |
|
|
|
| chr18_+_2928240 | 2.62 |
ENSDART00000165002
|
aqp11
|
aquaporin 11 |
| chr19_-_35642440 | 2.62 |
ENSDART00000167853
ENSDART00000054274 |
macf1a
|
microtubule-actin crosslinking factor 1a |
| chr20_+_47588954 | 2.59 |
ENSDART00000021341
|
kif3ca
|
kinesin family member 3Ca |
| chr9_+_39217373 | 2.53 |
ENSDART00000004742
|
cps1
|
carbamoyl-phosphate synthase 1, mitochondrial |
| KN150139v1_+_12765 | 2.48 |
|
|
|
| chr23_-_32378111 | 2.47 |
ENSDART00000143772
|
dgkaa
|
diacylglycerol kinase, alpha a |
| chr6_+_7309023 | 2.47 |
ENSDART00000160128
|
erbb3a
|
erb-b2 receptor tyrosine kinase 3a |
| chr11_+_34788205 | 2.43 |
ENSDART00000124800
|
fam212aa
|
family with sequence similarity 212, member Aa |
| chr20_-_3970778 | 2.43 |
ENSDART00000178724
ENSDART00000178565 |
TRIM67
|
tripartite motif containing 67 |
| chr3_+_23573114 | 2.40 |
ENSDART00000024256
|
hoxb6a
|
homeobox B6a |
| chr4_-_73618081 | 2.40 |
ENSDART00000157935
|
large
|
like-glycosyltransferase |
| chr9_-_12726136 | 2.38 |
|
|
|
| chr5_-_18895882 | 2.36 |
ENSDART00000008994
|
foxn4
|
forkhead box N4 |
| chr25_-_14953284 | 2.36 |
ENSDART00000165632
|
pax6a
|
paired box 6a |
| chr7_-_26331734 | 2.34 |
|
|
|
| chr5_-_69031855 | 2.32 |
ENSDART00000028954
|
rilpl2
|
Rab interacting lysosomal protein-like 2 |
| chr21_-_17566461 | 2.31 |
ENSDART00000044078
ENSDART00000168495 ENSDART00000020048 |
gsna
|
gelsolin a |
| chr5_+_32739678 | 2.31 |
ENSDART00000026085
|
ptges
|
prostaglandin E synthase |
| chr16_+_11260802 | 2.29 |
ENSDART00000140674
|
cicb
|
capicua transcriptional repressor b |
| chr22_-_36552513 | 2.29 |
ENSDART00000129318
|
CABZ01045212.1
|
ENSDARG00000087525 |
| chr23_-_10202175 | 2.29 |
ENSDART00000142442
|
plxnb1a
|
plexin b1a |
| chr8_+_15216833 | 2.28 |
ENSDART00000141185
|
slc5a9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr10_+_29378834 | 2.27 |
ENSDART00000155390
|
crebzf
|
CREB/ATF bZIP transcription factor |
| chr8_-_49443058 | 2.24 |
ENSDART00000011453
|
sypb
|
synaptophysin b |
| chr9_-_56877221 | 2.24 |
ENSDART00000168620
ENSDART00000171958 |
gpr39
|
G protein-coupled receptor 39 |
| chr11_-_39864306 | 2.20 |
ENSDART00000165781
|
fam83e
|
family with sequence similarity 83, member E |
| chr20_-_23127110 | 2.18 |
ENSDART00000015755
|
rasl11b
|
RAS-like, family 11, member B |
| chr2_-_43997672 | 2.17 |
ENSDART00000005449
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr7_-_16346150 | 2.16 |
ENSDART00000128488
|
e2f8
|
E2F transcription factor 8 |
| chr22_+_528076 | 2.16 |
|
|
|
| chr3_-_5318206 | 2.13 |
ENSDART00000137105
|
myh9b
|
myosin, heavy chain 9b, non-muscle |
| chr7_-_67906887 | 2.12 |
ENSDART00000171943
|
si:ch73-315f9.2
|
si:ch73-315f9.2 |
| chr9_+_50609926 | 2.11 |
ENSDART00000098687
|
GRB14
|
growth factor receptor bound protein 14 |
| chr13_-_22901327 | 2.09 |
ENSDART00000056523
|
hkdc1
|
hexokinase domain containing 1 |
| chr22_-_13018196 | 2.08 |
ENSDART00000028787
|
ahr1b
|
aryl hydrocarbon receptor 1b |
| chr9_-_24602136 | 2.07 |
ENSDART00000135897
|
tmeff2a
|
transmembrane protein with EGF-like and two follistatin-like domains 2a |
| chr5_-_65103664 | 2.05 |
ENSDART00000130888
|
notch1b
|
notch 1b |
| chr16_+_17706003 | 2.00 |
|
|
|
| chr18_+_23891152 | 1.99 |
ENSDART00000146490
|
BX569787.1
|
ENSDARG00000092402 |
| chr19_+_20164849 | 1.99 |
ENSDART00000169017
|
hoxa11a
|
homeobox A11a |
| chr25_+_7359375 | 1.93 |
|
|
|
| chr6_-_34024063 | 1.91 |
ENSDART00000003701
|
tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr2_+_45843528 | 1.90 |
ENSDART00000114225
ENSDART00000169279 |
CU467828.1
|
ENSDARG00000074082 |
| chr11_-_22200590 | 1.88 |
ENSDART00000006580
|
tfeb
|
transcription factor EB |
| chr11_-_39864543 | 1.86 |
ENSDART00000165781
|
fam83e
|
family with sequence similarity 83, member E |
| chr11_+_24052512 | 1.86 |
ENSDART00000029695
|
pnp4a
|
purine nucleoside phosphorylase 4a |
| chr20_-_8431338 | 1.85 |
ENSDART00000145841
ENSDART00000143936 ENSDART00000083906 ENSDART00000167102 ENSDART00000083908 |
dab1a
|
Dab, reelin signal transducer, homolog 1a (Drosophila) |
| chr2_-_26987186 | 1.84 |
ENSDART00000132854
|
u2surp
|
U2 snRNP-associated SURP domain containing |
| chr15_+_33212930 | 1.83 |
ENSDART00000164928
|
mab21l1
|
mab-21-like 1 |
| chr8_+_39525254 | 1.83 |
|
|
|
| chr13_+_21649237 | 1.81 |
ENSDART00000021556
|
si:ch211-51a6.2
|
si:ch211-51a6.2 |
| chr15_-_32738134 | 1.79 |
ENSDART00000164670
|
frem2b
|
Fras1 related extracellular matrix protein 2b |
| chr6_+_33556031 | 1.79 |
ENSDART00000147625
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr16_-_29502741 | 1.76 |
ENSDART00000148405
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr18_+_38768524 | 1.75 |
ENSDART00000143735
|
si:ch211-215d8.2
|
si:ch211-215d8.2 |
| chr16_+_23367625 | 1.75 |
ENSDART00000015956
|
efna1b
|
ephrin-A1b |
| chr23_+_25930072 | 1.75 |
ENSDART00000124103
|
hnf4a
|
hepatocyte nuclear factor 4, alpha |
| chr16_+_19731017 | 1.74 |
ENSDART00000139357
|
sp8b
|
sp8 transcription factor b |
| chr3_-_32409303 | 1.74 |
ENSDART00000151476
|
rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr8_-_16630507 | 1.74 |
|
|
|
| chr4_+_22749707 | 1.72 |
|
|
|
| chr8_+_19471914 | 1.71 |
ENSDART00000159044
|
sec22bb
|
SEC22 homolog B, vesicle trafficking protein b |
| chr4_-_76594399 | 1.70 |
ENSDART00000049170
|
zgc:85975
|
zgc:85975 |
| chr19_+_341775 | 1.68 |
ENSDART00000151013
|
ensaa
|
endosulfine alpha a |
| chr6_-_23455458 | 1.66 |
ENSDART00000157527
ENSDART00000168882 ENSDART00000171384 |
ntn1a
|
netrin 1a |
| chr8_-_52729505 | 1.65 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr10_+_3728747 | 1.64 |
|
|
|
| chr6_+_58704352 | 1.64 |
ENSDART00000172466
|
igsf8
|
immunoglobulin superfamily, member 8 |
| chr21_-_45573799 | 1.63 |
ENSDART00000158489
|
zgc:77058
|
zgc:77058 |
| chr6_+_41557636 | 1.63 |
ENSDART00000084834
|
srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr7_-_57030751 | 1.63 |
|
|
|
| chr21_+_9483423 | 1.62 |
ENSDART00000162834
|
mapk10
|
mitogen-activated protein kinase 10 |
| chr5_-_18896515 | 1.61 |
ENSDART00000008994
|
foxn4
|
forkhead box N4 |
| chr16_-_19385441 | 1.61 |
ENSDART00000118132
|
5S_rRNA
|
5S ribosomal RNA |
| chr12_-_28766713 | 1.61 |
ENSDART00000148459
|
cbx1b
|
chromobox homolog 1b (HP1 beta homolog Drosophila) |
| chr19_-_7621797 | 1.58 |
ENSDART00000143958
|
lix1l
|
limb and CNS expressed 1 like |
| chr22_-_22391686 | 1.58 |
ENSDART00000158555
|
camsap2a
|
calmodulin regulated spectrin-associated protein family, member 2a |
| chr15_-_18110169 | 1.58 |
|
|
|
| chr4_-_19992951 | 1.54 |
ENSDART00000169248
|
drd4-rs
|
dopamine receptor D4 related sequence |
| chr19_+_2291874 | 1.54 |
|
|
|
| chr21_+_37985816 | 1.54 |
ENSDART00000085728
|
klf8
|
Kruppel-like factor 8 |
| chr13_+_41885396 | 1.53 |
ENSDART00000131147
|
cyp1b1
|
cytochrome P450, family 1, subfamily B, polypeptide 1 |
| chr17_+_26946957 | 1.53 |
ENSDART00000114215
|
grhl3
|
grainyhead-like transcription factor 3 |
| chr23_-_2027166 | 1.53 |
ENSDART00000127443
|
prdm5
|
PR domain containing 5 |
| chr5_-_47520995 | 1.51 |
ENSDART00000144252
|
BX465834.1
|
ENSDARG00000095715 |
| chr21_+_26660833 | 1.50 |
ENSDART00000004109
|
gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr10_+_23052796 | 1.49 |
ENSDART00000138955
|
si:dkey-175g6.2
|
si:dkey-175g6.2 |
| chr19_-_9779467 | 1.49 |
|
|
|
| chr4_+_74835204 | 1.47 |
ENSDART00000172647
ENSDART00000174351 |
zgc:113209
|
zgc:113209 |
| chr4_-_29059042 | 1.47 |
ENSDART00000169971
|
CR387997.1
|
ENSDARG00000099486 |
| chr2_-_10264593 | 1.45 |
ENSDART00000091726
|
fam78ba
|
family with sequence similarity 78, member B a |
| chr3_-_48865399 | 1.44 |
ENSDART00000028610
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr14_-_29565869 | 1.44 |
ENSDART00000172349
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| KN150196v1_+_15525 | 1.44 |
|
|
|
| KN149817v1_+_2207 | 1.43 |
|
|
|
| chr18_+_23890892 | 1.42 |
ENSDART00000146490
|
BX569787.1
|
ENSDARG00000092402 |
| chr8_+_25984588 | 1.42 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 10.4 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 1.7 | 5.1 | GO:0010312 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 1.6 | 4.9 | GO:0072025 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 1.6 | 6.4 | GO:0021982 | pineal gland development(GO:0021982) |
| 1.5 | 4.6 | GO:0042756 | drinking behavior(GO:0042756) |
| 1.5 | 4.5 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) regulation of protein binding(GO:0043393) |
| 1.5 | 5.9 | GO:0061626 | ventral aorta development(GO:0035908) pharyngeal arch artery morphogenesis(GO:0061626) |
| 1.4 | 4.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 1.0 | 3.8 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.9 | 2.8 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 0.9 | 7.4 | GO:0060579 | ventral spinal cord interneuron fate commitment(GO:0060579) |
| 0.9 | 8.8 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.9 | 3.5 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.9 | 2.6 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.8 | 5.7 | GO:1902287 | semaphorin-plexin signaling pathway involved in neuron projection guidance(GO:1902285) semaphorin-plexin signaling pathway involved in axon guidance(GO:1902287) |
| 0.8 | 3.2 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.8 | 3.9 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.8 | 6.9 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.7 | 2.2 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.7 | 2.8 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.7 | 2.0 | GO:1903385 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.6 | 5.8 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.6 | 2.5 | GO:0019627 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.6 | 4.9 | GO:1904030 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.6 | 3.5 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.6 | 3.4 | GO:0060221 | retinal rod cell differentiation(GO:0060221) |
| 0.6 | 4.6 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.5 | 8.2 | GO:0035141 | medial fin morphogenesis(GO:0035141) |
| 0.5 | 1.5 | GO:0051350 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.5 | 3.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.5 | 3.5 | GO:1902975 | cell cycle DNA replication initiation(GO:1902292) nuclear cell cycle DNA replication initiation(GO:1902315) mitotic DNA replication initiation(GO:1902975) |
| 0.5 | 2.9 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.5 | 5.1 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.5 | 1.9 | GO:0097477 | spinal cord motor neuron migration(GO:0097476) lateral motor column neuron migration(GO:0097477) |
| 0.5 | 1.8 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.4 | 1.3 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.4 | 3.1 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.4 | 0.4 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.4 | 4.9 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.4 | 2.3 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.4 | 1.1 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.4 | 3.7 | GO:0072376 | protein activation cascade(GO:0072376) |
| 0.3 | 1.0 | GO:0044036 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.3 | 2.3 | GO:0001516 | prostaglandin biosynthetic process(GO:0001516) prostanoid biosynthetic process(GO:0046457) |
| 0.3 | 2.8 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.3 | 2.2 | GO:0048168 | regulation of neuronal synaptic plasticity(GO:0048168) |
| 0.3 | 2.5 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.3 | 2.1 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.2 | 5.2 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.2 | 1.2 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.2 | 0.7 | GO:0051311 | chiasma assembly(GO:0051026) meiotic metaphase plate congression(GO:0051311) |
| 0.2 | 2.9 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.2 | 1.1 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.2 | 0.8 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 | 1.4 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.2 | 1.7 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.2 | 2.7 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.2 | 2.8 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.2 | 3.5 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.2 | 1.3 | GO:0006833 | water transport(GO:0006833) |
| 0.2 | 4.7 | GO:0035113 | embryonic appendage morphogenesis(GO:0035113) |
| 0.2 | 1.9 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.2 | 2.1 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.2 | 1.9 | GO:0006959 | humoral immune response(GO:0006959) |
| 0.2 | 2.3 | GO:0003382 | epithelial cell morphogenesis(GO:0003382) |
| 0.2 | 1.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.2 | 1.4 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 0.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.7 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.1 | 0.7 | GO:0003173 | ventriculo bulbo valve development(GO:0003173) |
| 0.1 | 1.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.1 | 0.8 | GO:0019427 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.1 | 4.7 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.1 | 0.5 | GO:0042373 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) vitamin K metabolic process(GO:0042373) |
| 0.1 | 1.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.1 | 0.5 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 4.6 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.1 | 0.2 | GO:0060911 | cardiac cell fate commitment(GO:0060911) |
| 0.1 | 2.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 3.3 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.1 | 1.1 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 1.4 | GO:0072160 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) nephron tubule epithelial cell differentiation(GO:0072160) |
| 0.1 | 2.2 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.1 | 4.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.1 | 3.0 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.1 | 2.3 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 1.5 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) |
| 0.1 | 2.9 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 3.2 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.1 | 0.9 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 2.1 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.1 | 1.7 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 2.9 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.1 | 1.6 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 3.9 | GO:0031047 | gene silencing by RNA(GO:0031047) |
| 0.0 | 0.6 | GO:0030225 | macrophage differentiation(GO:0030225) |
| 0.0 | 0.1 | GO:0048714 | positive regulation of glial cell differentiation(GO:0045687) regulation of oligodendrocyte differentiation(GO:0048713) positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 4.8 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.1 | GO:0070285 | pigment cell development(GO:0070285) |
| 0.0 | 0.4 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 4.2 | GO:0060047 | heart contraction(GO:0060047) |
| 0.0 | 1.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 8.6 | GO:0050767 | regulation of neurogenesis(GO:0050767) |
| 0.0 | 1.4 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.7 | GO:0003146 | heart jogging(GO:0003146) |
| 0.0 | 1.6 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 2.3 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 2.3 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 1.2 | GO:1901653 | cellular response to peptide hormone stimulus(GO:0071375) cellular response to peptide(GO:1901653) |
| 0.0 | 1.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.1 | GO:0019229 | regulation of vasoconstriction(GO:0019229) |
| 0.0 | 1.1 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.5 | GO:0014020 | neural tube closure(GO:0001843) primary neural tube formation(GO:0014020) |
| 0.0 | 4.1 | GO:0006470 | protein dephosphorylation(GO:0006470) |
| 0.0 | 5.0 | GO:0006897 | endocytosis(GO:0006897) |
| 0.0 | 3.1 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 0.9 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 1.1 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.7 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 1.1 | GO:0030902 | hindbrain development(GO:0030902) |
| 0.0 | 0.8 | GO:0048232 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 0.5 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.0 | 0.8 | GO:0090630 | activation of GTPase activity(GO:0090630) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 9.0 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.8 | 5.7 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.8 | 3.2 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.7 | 2.8 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.7 | 2.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.7 | 2.6 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 0.6 | 4.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.2 | 3.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.2 | 0.7 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.2 | 2.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.2 | 2.9 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.2 | 1.1 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.2 | 3.5 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 1.6 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.2 | 4.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 7.5 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 5.8 | GO:0005604 | basement membrane(GO:0005604) |
| 0.1 | 1.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.1 | 3.0 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.1 | 0.6 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.1 | 2.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 2.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 5.1 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 2.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 2.9 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 4.9 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.1 | 2.7 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 3.4 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 2.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 7.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 23.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 1.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 3.1 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 2.6 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 2.1 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.9 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 6.9 | GO:0043005 | neuron projection(GO:0043005) |
| 0.0 | 5.6 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 4.2 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 8.4 | GO:1990904 | ribonucleoprotein complex(GO:1990904) |
| 0.0 | 1.7 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.6 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.6 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 1.0 | 3.0 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.0 | 3.9 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 1.0 | 3.9 | GO:0008158 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.9 | 2.8 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.8 | 2.5 | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.8 | 4.6 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.7 | 2.8 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.6 | 6.7 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.6 | 2.9 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.5 | 3.5 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.5 | 1.9 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.5 | 3.2 | GO:0015149 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.5 | 2.3 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.5 | 1.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.4 | 4.9 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.4 | 1.6 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.4 | 1.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.4 | 1.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.4 | 1.5 | GO:0099528 | G-protein coupled neurotransmitter receptor activity(GO:0099528) |
| 0.4 | 1.1 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.4 | 5.7 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.3 | 3.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.3 | 5.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.3 | 1.0 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.3 | 1.8 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.3 | 2.8 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.3 | 3.5 | GO:0043142 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.3 | 4.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 2.5 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 1.0 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.2 | 3.2 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.2 | 0.7 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.2 | 1.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 4.9 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.2 | 1.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 0.8 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.2 | 1.1 | GO:0008865 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 6.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.2 | 7.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 0.5 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.2 | 2.9 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 1.1 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 4.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.2 | 1.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 3.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 4.0 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 0.8 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.1 | 1.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.6 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.1 | 0.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 5.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.1 | 1.7 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 1.5 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.4 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.1 | 2.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.3 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.1 | 2.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 1.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 5.5 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 2.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 0.6 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 1.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 65.5 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 2.9 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 2.5 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 2.0 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 2.4 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 3.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 2.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.6 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 1.4 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 5.2 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 0.1 | GO:0008311 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 4.2 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 14.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 9.0 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 1.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.2 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 4.2 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 2.2 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 1.8 | GO:0016779 | nucleotidyltransferase activity(GO:0016779) |
| 0.0 | 0.6 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.0 | GO:0039706 | co-receptor binding(GO:0039706) receptor antagonist activity(GO:0048019) |
| 0.0 | 1.5 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.2 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 0.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 6.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 1.8 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.2 | 7.4 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.2 | 3.9 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 2.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 4.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 0.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 2.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.1 | 1.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 3.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 2.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.1 | 0.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 3.4 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 1.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 0.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 3.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.9 | 5.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.7 | 3.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.5 | 7.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.3 | 4.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.3 | 7.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.3 | 2.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.2 | 3.5 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 6.2 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.2 | 3.4 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 2.6 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.2 | 1.6 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 5.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 1.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.2 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 0.5 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.2 | 6.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 1.3 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 2.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.1 | 0.4 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.1 | 3.1 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.1 | 2.9 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 2.2 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.1 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 1.4 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.1 | 1.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 1.1 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 1.2 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 2.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.6 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.7 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.1 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |