Project
DANIO-CODE
Navigation
Downloads
Results for si:dkey-68o6.8
Z-value: 0.34
Transcription factors associated with si:dkey-68o6.8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
si_dkey-68o6.8
|
ENSDARG00000104144 | si_dkey-68o6.8 |
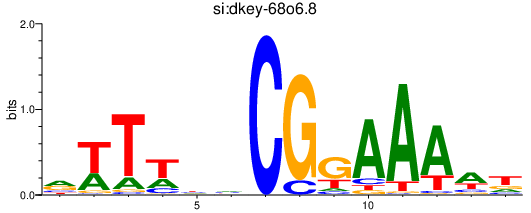
Activity profile of si:dkey-68o6.8 motif
Sorted Z-values of si:dkey-68o6.8 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of si:dkey-68o6.8
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_26333957 | 1.12 |
ENSDART00000130493
|
capn2b
|
calpain 2, (m/II) large subunit b |
| chr13_-_5440923 | 1.04 |
ENSDART00000102576
|
meis1b
|
Meis homeobox 1 b |
| chr24_-_37680649 | 1.03 |
ENSDART00000056286
|
h1f0
|
H1 histone family, member 0 |
| chr3_+_24067387 | 0.77 |
ENSDART00000055609
|
atf4b
|
activating transcription factor 4b |
| chr4_+_22959458 | 0.72 |
ENSDART00000036531
|
gnai1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr11_-_15162109 | 0.65 |
ENSDART00000124968
|
rpn2
|
ribophorin II |
| chr3_+_18249107 | 0.64 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr23_-_28420470 | 0.63 |
ENSDART00000145072
|
neurod4
|
neuronal differentiation 4 |
| chr2_-_23516930 | 0.61 |
ENSDART00000165355
|
prrx1a
|
paired related homeobox 1a |
| chr4_+_5515028 | 0.54 |
ENSDART00000027304
|
mapk12b
|
mitogen-activated protein kinase 12b |
| chr14_-_17258072 | 0.54 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor-like 1a |
| chr8_-_32795895 | 0.53 |
ENSDART00000131597
|
zgc:194839
|
zgc:194839 |
| chr6_+_54231519 | 0.51 |
ENSDART00000149542
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr20_-_7303735 | 0.51 |
ENSDART00000100060
|
dsc2l
|
desmocollin 2 like |
| chr24_-_26334717 | 0.51 |
ENSDART00000079984
ENSDART00000136871 |
rpl22l1
|
ribosomal protein L22-like 1 |
| chr11_-_40416837 | 0.49 |
ENSDART00000166372
|
si:ch211-222l21.1
|
si:ch211-222l21.1 |
| chr15_+_44183291 | 0.46 |
ENSDART00000168515
|
CU929391.1
|
ENSDARG00000088039 |
| chr2_-_23517033 | 0.45 |
ENSDART00000041365
|
prrx1a
|
paired related homeobox 1a |
| chr1_+_50835265 | 0.43 |
ENSDART00000162226
|
meis1a
|
Meis homeobox 1 a |
| chr12_-_3018686 | 0.43 |
ENSDART00000169161
|
dcxr
|
dicarbonyl/L-xylulose reductase |
| chr7_-_35161302 | 0.43 |
ENSDART00000026712
|
mmp2
|
matrix metallopeptidase 2 |
| chr10_-_11806373 | 0.41 |
ENSDART00000009715
|
adamts6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr14_-_17257773 | 0.41 |
ENSDART00000082667
|
fgfrl1a
|
fibroblast growth factor receptor-like 1a |
| chr18_+_45199548 | 0.41 |
|
|
|
| chr7_+_13753344 | 0.41 |
|
|
|
| chr5_+_49093134 | 0.40 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr24_-_25099449 | 0.40 |
ENSDART00000153798
|
hhla2b.2
|
HERV-H LTR-associating 2b, tandem duplicate 2 |
| chr6_-_9346394 | 0.39 |
ENSDART00000144335
|
cyp27c1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr22_-_38621100 | 0.39 |
ENSDART00000164609
|
si:ch211-126j24.1
|
si:ch211-126j24.1 |
| chr7_-_52142689 | 0.38 |
ENSDART00000110265
|
myzap
|
myocardial zonula adherens protein |
| chr18_-_14723138 | 0.37 |
ENSDART00000010129
|
pdf
|
peptide deformylase (mitochondrial) |
| chr5_+_37861949 | 0.37 |
ENSDART00000144425
|
gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr18_-_605558 | 0.34 |
ENSDART00000157564
|
wtip
|
WT1 interacting protein |
| chr5_-_36349284 | 0.34 |
ENSDART00000047269
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr20_-_7303804 | 0.33 |
ENSDART00000100060
|
dsc2l
|
desmocollin 2 like |
| chr8_+_21321765 | 0.32 |
ENSDART00000131691
|
alas2
|
aminolevulinate, delta-, synthase 2 |
| chr14_+_6834563 | 0.31 |
ENSDART00000170994
ENSDART00000129898 |
ctsf
|
cathepsin F |
| chr6_-_41087828 | 0.31 |
ENSDART00000028217
|
srsf3a
|
serine/arginine-rich splicing factor 3a |
| chr10_-_10374032 | 0.30 |
ENSDART00000134929
|
BX323065.1
|
ENSDARG00000095365 |
| chr17_+_29328653 | 0.30 |
ENSDART00000086164
|
kctd3
|
potassium channel tetramerization domain containing 3 |
| chr22_-_31722970 | 0.29 |
ENSDART00000128247
|
lsm3
|
LSM3 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr17_+_25407644 | 0.29 |
ENSDART00000149557
|
srrm1
|
serine/arginine repetitive matrix 1 |
| chr5_+_15880920 | 0.28 |
ENSDART00000168643
|
CABZ01088700.1
|
ENSDARG00000101452 |
| chr19_+_46647893 | 0.28 |
ENSDART00000162785
ENSDART00000171251 ENSDART00000164938 |
mapk15
|
mitogen-activated protein kinase 15 |
| chr15_-_47234665 | 0.28 |
ENSDART00000124474
|
CU633855.1
|
ENSDARG00000089563 |
| chr20_+_37480145 | 0.28 |
ENSDART00000134997
ENSDART00000145927 |
adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr9_-_695593 | 0.28 |
ENSDART00000115030
|
dip2a
|
disco-interacting protein 2 homolog A |
| chr4_+_46379457 | 0.27 |
ENSDART00000162948
|
BX324155.1
|
ENSDARG00000104638 |
| chr16_-_4811237 | 0.27 |
ENSDART00000076955
|
cyp4t8
|
cytochrome P450, family 4, subfamily T, polypeptide 8 |
| chr1_+_35058017 | 0.27 |
ENSDART00000085051
|
hhip
|
hedgehog interacting protein |
| chr6_+_53349584 | 0.26 |
ENSDART00000167079
|
si:ch211-161c3.5
|
si:ch211-161c3.5 |
| chr2_+_25468904 | 0.26 |
ENSDART00000056801
|
msl2a
|
male-specific lethal 2 homolog a (Drosophila) |
| chr15_+_44216276 | 0.26 |
|
|
|
| chr22_-_10372023 | 0.25 |
|
|
|
| chr7_-_72127164 | 0.25 |
ENSDART00000176069
|
CABZ01057364.1
|
ENSDARG00000106736 |
| chr9_-_10561062 | 0.24 |
ENSDART00000175269
|
thsd7ba
|
thrombospondin, type I, domain containing 7Ba |
| chr6_+_59749159 | 0.24 |
|
|
|
| chr16_-_6450531 | 0.23 |
ENSDART00000060550
ENSDART00000145663 |
ndufb9
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9 |
| chr11_+_11809234 | 0.23 |
ENSDART00000168346
|
CR848705.1
|
ENSDARG00000102054 |
| chr7_+_73598802 | 0.23 |
ENSDART00000109720
|
zgc:163061
|
zgc:163061 |
| chr2_+_54665352 | 0.23 |
|
|
|
| chr17_+_45832040 | 0.23 |
ENSDART00000154638
|
kif26ab
|
kinesin family member 26Ab |
| chr10_-_42258801 | 0.22 |
ENSDART00000063071
|
dgcr2
|
DiGeorge syndrome critical region gene 2 |
| chr21_-_38571536 | 0.21 |
ENSDART00000139178
ENSDART00000036600 |
slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr2_+_56935721 | 0.20 |
ENSDART00000168497
|
eef2b
|
eukaryotic translation elongation factor 2b |
| chr22_-_36561247 | 0.20 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr13_+_28382422 | 0.20 |
ENSDART00000043117
|
fbxw4
|
F-box and WD repeat domain containing 4 |
| chr6_-_20625494 | 0.20 |
|
|
|
| chr5_+_52934865 | 0.19 |
ENSDART00000161682
|
BX323994.2
|
ENSDARG00000100423 |
| chr2_-_13322929 | 0.18 |
|
|
|
| chr23_-_41929027 | 0.18 |
ENSDART00000131235
|
GMEB2
|
glucocorticoid modulatory element binding protein 2 |
| chr19_-_6466494 | 0.17 |
ENSDART00000104950
|
atp1a3a
|
ATPase, Na+/K+ transporting, alpha 3a polypeptide |
| chr4_+_5515454 | 0.17 |
ENSDART00000027304
|
mapk12b
|
mitogen-activated protein kinase 12b |
| chr11_-_40416762 | 0.17 |
ENSDART00000166372
|
si:ch211-222l21.1
|
si:ch211-222l21.1 |
| chr21_+_35292807 | 0.17 |
ENSDART00000130626
|
rars
|
arginyl-tRNA synthetase |
| chr22_+_1360497 | 0.17 |
ENSDART00000160330
|
BX322657.1
|
ENSDARG00000102448 |
| chr6_-_32000976 | 0.17 |
ENSDART00000132280
|
ror1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr16_+_41862759 | 0.16 |
|
|
|
| chr8_-_13934852 | 0.16 |
ENSDART00000133830
|
serping1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr10_+_45213089 | 0.16 |
ENSDART00000166945
|
nudcd3
|
NudC domain containing 3 |
| chr21_+_37751640 | 0.16 |
ENSDART00000076328
|
pgrmc1
|
progesterone receptor membrane component 1 |
| chr14_-_27703812 | 0.16 |
ENSDART00000054115
|
tsc22d3
|
TSC22 domain family, member 3 |
| chr20_+_27048717 | 0.15 |
ENSDART00000062066
|
si:dkey-177p2.6
|
si:dkey-177p2.6 |
| chr13_+_16148867 | 0.15 |
ENSDART00000142408
|
anxa11a
|
annexin A11a |
| chr13_-_29904289 | 0.14 |
ENSDART00000143745
ENSDART00000041702 |
trmt2b
|
tRNA methyltransferase 2 homolog B |
| chr16_-_43029548 | 0.14 |
ENSDART00000057305
|
thbs3a
|
thrombospondin 3a |
| chr4_+_3442322 | 0.14 |
ENSDART00000058277
|
znf800b
|
zinc finger protein 800b |
| chr6_+_11770822 | 0.14 |
ENSDART00000128024
ENSDART00000121431 |
wdsub1
|
WD repeat, sterile alpha motif and U-box domain containing 1 |
| chr20_-_7303680 | 0.14 |
ENSDART00000100060
|
dsc2l
|
desmocollin 2 like |
| chr24_-_6346381 | 0.14 |
ENSDART00000013294
|
anxa13
|
annexin A13 |
| chr11_-_36017515 | 0.14 |
ENSDART00000122531
|
nfya
|
nuclear transcription factor Y, alpha |
| chr10_-_13281047 | 0.13 |
ENSDART00000001253
|
si:busm1-57f23.1
|
si:busm1-57f23.1 |
| chr8_-_46378116 | 0.13 |
ENSDART00000136602
|
qars
|
glutaminyl-tRNA synthetase |
| chr11_-_36017572 | 0.13 |
ENSDART00000099571
|
nfya
|
nuclear transcription factor Y, alpha |
| chr21_-_5692160 | 0.13 |
ENSDART00000075137
ENSDART00000151202 |
ccni
|
cyclin I |
| chr23_-_27930478 | 0.13 |
ENSDART00000043462
|
acvrl1
|
activin A receptor type II-like 1 |
| chr4_-_5294280 | 0.13 |
ENSDART00000178921
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr15_-_33688186 | 0.13 |
|
|
|
| chr21_-_2247380 | 0.12 |
ENSDART00000171417
|
si:ch73-299h12.3
|
si:ch73-299h12.3 |
| chr1_-_11474269 | 0.12 |
ENSDART00000054801
|
b4galt1
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
| chr3_+_18249890 | 0.12 |
ENSDART00000110842
|
rps2
|
ribosomal protein S2 |
| chr8_-_11132386 | 0.12 |
ENSDART00000133532
|
ENSDARG00000078221
|
ENSDARG00000078221 |
| chr2_+_26184009 | 0.11 |
ENSDART00000134077
|
eif5a2
|
eukaryotic translation initiation factor 5A2 |
| chr13_+_1227220 | 0.11 |
ENSDART00000114907
|
ifngr1
|
interferon gamma receptor 1 |
| chr2_+_25037678 | 0.11 |
ENSDART00000170467
|
pik3r2
|
phosphoinositide-3-kinase, regulatory subunit 2 (beta) |
| chr19_+_20156598 | 0.11 |
ENSDART00000171610
|
hoxa13a
|
homeobox A13a |
| chr19_-_32913075 | 0.11 |
ENSDART00000052104
|
fuca1.1
|
fucosidase, alpha-L- 1, tissue, tandem duplicate 1 |
| chr19_+_46631965 | 0.11 |
ENSDART00000158703
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr2_-_23516991 | 0.11 |
ENSDART00000165355
|
prrx1a
|
paired related homeobox 1a |
| chr24_-_26339981 | 0.10 |
ENSDART00000135496
|
eif5a
|
eukaryotic translation initiation factor 5A |
| chr1_+_585442 | 0.10 |
ENSDART00000147633
|
mrpl39
|
mitochondrial ribosomal protein L39 |
| chr9_-_41072329 | 0.10 |
ENSDART00000066424
|
pofut2
|
protein O-fucosyltransferase 2 |
| chr6_+_40664212 | 0.10 |
ENSDART00000103842
|
eno1b
|
enolase 1b, (alpha) |
| chr2_-_58737903 | 0.10 |
|
|
|
| chr17_-_43526955 | 0.10 |
ENSDART00000133665
|
mrpl35
|
mitochondrial ribosomal protein L35 |
| chr22_-_2942708 | 0.09 |
ENSDART00000040701
|
ing5a
|
inhibitor of growth family, member 5a |
| chr23_-_18646217 | 0.09 |
ENSDART00000138599
|
ssr4
|
signal sequence receptor, delta |
| chr7_+_22538681 | 0.09 |
ENSDART00000020212
|
sf1
|
splicing factor 1 |
| chr11_+_13000957 | 0.09 |
ENSDART00000161532
|
zfyve9b
|
zinc finger, FYVE domain containing 9b |
| chr15_-_44093289 | 0.09 |
ENSDART00000078086
|
CU655961.1
|
ENSDARG00000055690 |
| chr7_+_22539037 | 0.09 |
ENSDART00000020212
|
sf1
|
splicing factor 1 |
| chr18_+_14627030 | 0.09 |
ENSDART00000080788
|
wfdc1
|
WAP four-disulfide core domain 1 |
| chr11_+_2527448 | 0.09 |
ENSDART00000175330
|
dnajc14
|
DnaJ (Hsp40) homolog, subfamily C, member 14 |
| chr17_+_11904378 | 0.09 |
ENSDART00000150209
ENSDART00000110058 |
tfb2m
|
transcription factor B2, mitochondrial |
| chr24_-_18775194 | 0.09 |
ENSDART00000106188
|
cpa6
|
carboxypeptidase A6 |
| chr4_+_76581339 | 0.09 |
ENSDART00000174203
|
zgc:113921
|
zgc:113921 |
| chr2_-_27920046 | 0.09 |
ENSDART00000014568
|
urod
|
uroporphyrinogen decarboxylase |
| chr8_-_11164125 | 0.08 |
ENSDART00000147817
|
fam208b
|
family with sequence similarity 208, member B |
| chr14_-_529171 | 0.08 |
ENSDART00000171317
|
SPATA5
|
spermatogenesis associated 5 |
| chr3_+_24067430 | 0.08 |
ENSDART00000055609
|
atf4b
|
activating transcription factor 4b |
| chr3_+_40173549 | 0.08 |
ENSDART00000083241
|
slc29a4
|
solute carrier family 29 (equilibrative nucleoside transporter), member 4 |
| chr19_-_19931617 | 0.08 |
ENSDART00000160582
|
creb5a
|
cAMP responsive element binding protein 5a |
| chr14_-_51717514 | 0.08 |
ENSDART00000164272
|
rest
|
RE1-silencing transcription factor |
| chr1_-_11474192 | 0.08 |
ENSDART00000054801
|
b4galt1
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
| chr9_+_41072345 | 0.08 |
ENSDART00000141548
|
hibch
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr19_-_43434820 | 0.07 |
ENSDART00000122135
|
zcchc17
|
zinc finger, CCHC domain containing 17 |
| chr15_-_1070691 | 0.07 |
ENSDART00000153569
|
si:dkey-77f5.4
|
si:dkey-77f5.4 |
| chr3_-_50386208 | 0.07 |
ENSDART00000156709
|
BX088529.1
|
ENSDARG00000097178 |
| chr1_+_44301612 | 0.07 |
ENSDART00000021336
|
dnaja1
|
DnaJ (Hsp40) homolog, subfamily A, member 1 |
| chr3_-_18234622 | 0.06 |
ENSDART00000027630
|
kdelr2a
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2a |
| chr22_+_31866783 | 0.06 |
ENSDART00000159825
|
dock3
|
dedicator of cytokinesis 3 |
| chr21_-_33961137 | 0.06 |
ENSDART00000100508
|
ebf1b
|
early B-cell factor 1b |
| chr1_-_22583123 | 0.06 |
ENSDART00000122877
|
rpl9
|
ribosomal protein L9 |
| chr19_-_8617513 | 0.06 |
ENSDART00000139715
|
dpm3
|
dolichyl-phosphate mannosyltransferase polypeptide 3 |
| chr1_+_50348973 | 0.06 |
|
|
|
| chr10_+_45213266 | 0.06 |
ENSDART00000166945
|
nudcd3
|
NudC domain containing 3 |
| chr13_+_33173742 | 0.05 |
ENSDART00000145295
|
dcdc2b
|
doublecortin domain containing 2B |
| chr25_+_3884499 | 0.05 |
ENSDART00000104926
|
ENSDARG00000058108
|
ENSDARG00000058108 |
| chr12_-_33871785 | 0.05 |
ENSDART00000105545
|
arl3
|
ADP-ribosylation factor-like 3 |
| chr8_+_20830940 | 0.05 |
ENSDART00000015891
|
fzr1a
|
fizzy/cell division cycle 20 related 1a |
| chr2_+_22708278 | 0.05 |
ENSDART00000163172
|
hs2st1a
|
heparan sulfate 2-O-sulfotransferase 1a |
| chr1_-_8293777 | 0.05 |
ENSDART00000133496
|
CU467110.1
|
ENSDARG00000093438 |
| chr18_-_21229799 | 0.04 |
ENSDART00000060160
|
calb2a
|
calbindin 2a |
| chr2_-_54028761 | 0.04 |
ENSDART00000178289
|
CABZ01050256.1
|
ENSDARG00000109075 |
| chr7_+_13750087 | 0.04 |
ENSDART00000091470
|
furina
|
furin (paired basic amino acid cleaving enzyme) a |
| chr1_+_52157637 | 0.04 |
ENSDART00000123972
|
smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 |
| chr9_-_48152388 | 0.04 |
|
|
|
| chr9_+_6600592 | 0.04 |
ENSDART00000061577
|
fhl2a
|
four and a half LIM domains 2a |
| chr7_-_65909983 | 0.04 |
ENSDART00000154961
|
btbd10b
|
BTB (POZ) domain containing 10b |
| chr4_-_18617553 | 0.04 |
ENSDART00000171450
ENSDART00000172976 |
CDPF1
|
cysteine rich DPF motif domain containing 1 |
| chr7_+_26195619 | 0.04 |
ENSDART00000058908
|
mpdu1b
|
mannose-P-dolichol utilization defect 1b |
| chr3_+_41584507 | 0.03 |
ENSDART00000154401
|
chst12a
|
carbohydrate (chondroitin 4) sulfotransferase 12a |
| chr18_+_17032264 | 0.03 |
ENSDART00000100160
|
chtf8
|
CTF8, chromosome transmission fidelity factor 8 homolog (S. cerevisiae) |
| chr6_+_13273286 | 0.03 |
ENSDART00000032331
|
gmppab
|
GDP-mannose pyrophosphorylase Ab |
| chr19_+_43434941 | 0.03 |
ENSDART00000038230
|
snrnp40
|
small nuclear ribonucleoprotein 40 (U5) |
| chr19_-_6963103 | 0.03 |
|
|
|
| chr9_-_53138743 | 0.03 |
ENSDART00000085284
|
pfkla
|
phosphofructokinase, liver a |
| chr4_-_5293599 | 0.03 |
ENSDART00000177099
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr12_+_18914438 | 0.03 |
ENSDART00000153086
|
kctd17
|
potassium channel tetramerization domain containing 17 |
| chr1_-_11191948 | 0.02 |
ENSDART00000163971
ENSDART00000123431 |
iqce
|
IQ motif containing E |
| chr9_-_34410053 | 0.02 |
ENSDART00000111027
|
dcaf6
|
ddb1 and cul4 associated factor 6 |
| chr3_+_30927580 | 0.02 |
ENSDART00000153381
|
si:dkey-66i24.7
|
si:dkey-66i24.7 |
| chr19_+_7235553 | 0.01 |
ENSDART00000160894
|
brd2a
|
bromodomain containing 2a |
| chr9_-_29850713 | 0.01 |
ENSDART00000156609
|
CU855930.2
|
ENSDARG00000097441 |
| chr19_-_46631565 | 0.01 |
|
|
|
| chr17_-_21773194 | 0.01 |
ENSDART00000160296
|
hmx3a
|
H6 family homeobox 3a |
| chr17_-_17928974 | 0.01 |
ENSDART00000090447
|
hhipl1
|
HHIP-like 1 |
| chr13_+_33173790 | 0.01 |
ENSDART00000145295
|
dcdc2b
|
doublecortin domain containing 2B |
| chr10_+_25234636 | 0.00 |
ENSDART00000024514
|
chordc1b
|
cysteine and histidine-rich domain (CHORD) containing 1b |
| chr12_+_48176075 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0086014 | atrial cardiac muscle cell action potential(GO:0086014) cell-cell signaling involved in cardiac conduction(GO:0086019) atrial cardiac muscle cell to AV node cell signaling(GO:0086026) cell communication involved in cardiac conduction(GO:0086065) atrial cardiac muscle cell to AV node cell communication(GO:0086066) |
| 0.1 | 0.4 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.1 | 0.4 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.1 | 1.0 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.9 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.1 | 0.3 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.1 | 0.2 | GO:1903429 | regulation of cell maturation(GO:1903429) |
| 0.1 | 0.5 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.2 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.1 | 1.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.1 | 0.3 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.1 | 0.6 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.1 | 0.4 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.3 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.4 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.0 | 0.2 | GO:0015744 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) malate transmembrane transport(GO:0071423) |
| 0.0 | 0.4 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 0.0 | 0.3 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.4 | GO:0046501 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.2 | GO:0002526 | acute inflammatory response(GO:0002526) |
| 0.0 | 0.1 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 0.1 | GO:0060231 | mesenchymal to epithelial transition(GO:0060231) |
| 0.0 | 0.2 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.3 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.1 | GO:0034405 | response to fluid shear stress(GO:0034405) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.0 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.0 | 0.2 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 0.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.3 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.8 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.8 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.1 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.0 | 0.7 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.3 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase II, core complex(GO:0005665) DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0019841 | retinol binding(GO:0019841) |
| 0.1 | 0.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 1.0 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.2 | GO:0015140 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.7 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 0.1 | GO:0015928 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 2.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0046922 | peptide-O-fucosyltransferase activity(GO:0046922) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.2 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.0 | GO:1990757 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.0 | 0.3 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.4 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.0 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 0.7 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.3 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |