Project
DANIO-CODE
Navigation
Downloads
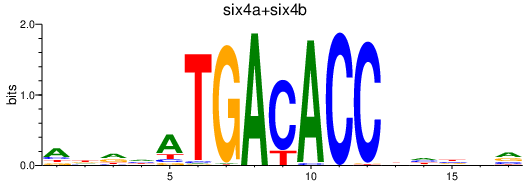
Results for six4a+six4b
Z-value: 0.88
Transcription factors associated with six4a+six4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
six4a
|
ENSDARG00000004695 | SIX homeobox 4a |
|
six4b
|
ENSDARG00000031983 | SIX homeobox 4b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| six4a | dr10_dc_chr13_-_31516399_31516443 | 0.77 | 5.5e-04 | Click! |
| six4b | dr10_dc_chr20_+_20585136_20585215 | 0.67 | 4.2e-03 | Click! |
Activity profile of six4a+six4b motif
Sorted Z-values of six4a+six4b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of six4a+six4b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_11113032 | 2.76 |
ENSDART00000027598
|
tpm3
|
tropomyosin 3 |
| chr7_-_26035308 | 2.18 |
ENSDART00000131906
|
zgc:77439
|
zgc:77439 |
| chr2_+_24649130 | 2.17 |
ENSDART00000078972
|
fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr23_+_36002332 | 2.01 |
ENSDART00000103139
|
hoxc8a
|
homeobox C8a |
| chr16_-_17439735 | 1.61 |
ENSDART00000144392
|
zyx
|
zyxin |
| chr21_-_43020159 | 1.57 |
ENSDART00000065097
|
dpysl3
|
dihydropyrimidinase-like 3 |
| chr24_+_36045759 | 1.46 |
ENSDART00000173322
|
obscnb
|
obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF b |
| chr4_-_16356071 | 1.41 |
ENSDART00000079521
|
kera
|
keratocan |
| chr13_-_22877318 | 1.39 |
ENSDART00000057638
ENSDART00000171778 |
hk1
|
hexokinase 1 |
| KN150642v1_+_9419 | 1.35 |
ENSDART00000159069
|
atoh1b
|
atonal bHLH transcription factor 1b |
| chr2_+_33352680 | 1.34 |
ENSDART00000126135
|
slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr2_+_43619752 | 1.25 |
ENSDART00000142078
ENSDART00000098265 ENSDART00000098267 |
nrp1b
|
neuropilin 1b |
| chr12_+_22605268 | 1.24 |
|
|
|
| chr2_+_11973282 | 1.20 |
ENSDART00000057268
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr23_-_19898781 | 1.19 |
ENSDART00000144027
ENSDART00000139425 |
haus7
|
HAUS augmin-like complex, subunit 7 |
| chr17_+_7438152 | 1.16 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr19_+_10412345 | 1.15 |
ENSDART00000143930
|
tmem238a
|
transmembrane protein 238a |
| chr11_+_15935769 | 1.12 |
ENSDART00000158824
|
ctbs
|
chitobiase, di-N-acetyl- |
| chr10_-_44509732 | 1.10 |
|
|
|
| chr16_-_24646282 | 1.09 |
ENSDART00000126274
|
si:ch211-79k12.2
|
si:ch211-79k12.2 |
| chr23_+_2597825 | 1.07 |
ENSDART00000159039
|
ENSDARG00000089986
|
ENSDARG00000089986 |
| chr14_-_6822151 | 1.04 |
ENSDART00000171311
|
si:ch73-43g23.1
|
si:ch73-43g23.1 |
| chr9_+_27939731 | 1.02 |
ENSDART00000125303
|
rab5b
|
RAB5B, member RAS oncogene family |
| chr16_+_21436660 | 1.00 |
ENSDART00000145886
|
osbpl3b
|
oxysterol binding protein-like 3b |
| chr2_+_29507819 | 0.99 |
|
|
|
| chr10_+_44509370 | 0.96 |
|
|
|
| chr11_+_40385236 | 0.94 |
ENSDART00000173156
|
slc45a1
|
solute carrier family 45, member 1 |
| chr8_+_49582361 | 0.93 |
ENSDART00000108613
|
rasef
|
RAS and EF-hand domain containing |
| chr7_+_58397256 | 0.93 |
ENSDART00000049264
|
sdr16c5b
|
short chain dehydrogenase/reductase family 16C, member 5b |
| chr10_+_33956052 | 0.90 |
|
|
|
| chr2_-_11879076 | 0.86 |
ENSDART00000145108
|
ENSDARG00000039203
|
ENSDARG00000039203 |
| chr2_+_50892294 | 0.86 |
ENSDART00000018150
ENSDART00000111135 |
neurod6b
|
neuronal differentiation 6b |
| chr12_+_22605229 | 0.85 |
|
|
|
| chr7_-_26035541 | 0.84 |
ENSDART00000057288
|
zgc:77439
|
zgc:77439 |
| chr10_+_12559022 | 0.81 |
ENSDART00000159853
|
FQ311890.1
|
ENSDARG00000102794 |
| chr2_-_32575527 | 0.81 |
ENSDART00000140026
|
smarcd3a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3a |
| chr2_-_40170303 | 0.80 |
ENSDART00000165602
|
epha4a
|
eph receptor A4a |
| chr16_+_14135089 | 0.79 |
|
|
|
| chr17_-_31041977 | 0.79 |
ENSDART00000104307
|
eml1
|
echinoderm microtubule associated protein like 1 |
| chr6_-_40660116 | 0.71 |
ENSDART00000154359
|
ppil1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr22_+_34454253 | 0.71 |
ENSDART00000156615
|
amigo3
|
adhesion molecule with Ig-like domain 3 |
| chr25_-_15117843 | 0.69 |
ENSDART00000031499
|
wt1a
|
wilms tumor 1a |
| chr1_+_37033111 | 0.68 |
ENSDART00000139448
|
GALNTL6
|
polypeptide N-acetylgalactosaminyltransferase like 6 |
| chr2_-_30787161 | 0.66 |
ENSDART00000131230
|
rgs20
|
regulator of G protein signaling 20 |
| chr3_-_54352640 | 0.66 |
ENSDART00000128041
|
dnmt1
|
DNA (cytosine-5-)-methyltransferase 1 |
| chr20_+_27792255 | 0.65 |
ENSDART00000153299
|
si:dkey-1h6.8
|
si:dkey-1h6.8 |
| chr3_-_35425227 | 0.61 |
ENSDART00000010944
|
dctn5
|
dynactin 5 |
| chr14_-_40454194 | 0.60 |
ENSDART00000166621
|
ef1
|
E74-like factor 1 (ets domain transcription factor) |
| chr8_-_43938411 | 0.59 |
|
|
|
| chr3_+_18249890 | 0.59 |
ENSDART00000110842
|
rps2
|
ribosomal protein S2 |
| chr6_-_35326559 | 0.59 |
ENSDART00000114960
|
nos1apa
|
nitric oxide synthase 1 (neuronal) adaptor protein a |
| chr21_+_29190291 | 0.56 |
ENSDART00000013641
|
adra1ba
|
adrenoceptor alpha 1Ba |
| chr22_+_4750764 | 0.52 |
ENSDART00000106156
|
mydgf
|
myeloid-derived growth factor |
| chr2_-_21008962 | 0.51 |
ENSDART00000114546
|
dusp12
|
dual specificity phosphatase 12 |
| chr4_-_71046688 | 0.51 |
ENSDART00000161800
|
CABZ01054391.1
|
ENSDARG00000104152 |
| chr10_-_36738730 | 0.50 |
ENSDART00000139097
|
dnajb13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr10_+_248929 | 0.50 |
|
|
|
| chr7_+_73604595 | 0.49 |
ENSDART00000109316
|
zgc:173587
|
zgc:173587 |
| chr16_+_14135138 | 0.48 |
|
|
|
| chr6_+_49584462 | 0.46 |
ENSDART00000103357
|
vapb
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr9_+_48521741 | 0.44 |
ENSDART00000145972
|
ccdc173
|
coiled-coil domain containing 173 |
| chr2_+_43619953 | 0.43 |
ENSDART00000142078
ENSDART00000098265 ENSDART00000098267 |
nrp1b
|
neuropilin 1b |
| chr15_+_11798993 | 0.40 |
ENSDART00000163844
|
si:dkey-31c13.1
|
si:dkey-31c13.1 |
| chr17_-_42778156 | 0.37 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr2_+_43620000 | 0.36 |
ENSDART00000142078
ENSDART00000098265 ENSDART00000098267 |
nrp1b
|
neuropilin 1b |
| chr6_+_13273286 | 0.36 |
ENSDART00000032331
|
gmppab
|
GDP-mannose pyrophosphorylase Ab |
| chr13_-_5423165 | 0.35 |
|
|
|
| chr15_+_17924364 | 0.35 |
|
|
|
| chr18_+_19430014 | 0.33 |
ENSDART00000025107
|
map2k1
|
mitogen-activated protein kinase kinase 1 |
| chr25_-_20835675 | 0.32 |
|
|
|
| chr9_+_41072345 | 0.32 |
ENSDART00000141548
|
hibch
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr4_-_22798521 | 0.29 |
ENSDART00000002851
ENSDART00000123801 ENSDART00000130409 |
kdm7aa
|
lysine (K)-specific demethylase 7Aa |
| chr7_+_39089755 | 0.28 |
ENSDART00000173481
|
acp2
|
acid phosphatase 2, lysosomal |
| chr18_-_26912259 | 0.25 |
ENSDART00000147735
|
si:dkey-24l11.2
|
si:dkey-24l11.2 |
| chr2_-_43886373 | 0.24 |
ENSDART00000138947
|
kif5ba
|
kinesin family member 5B, a |
| chr20_+_26196386 | 0.23 |
ENSDART00000139350
|
syne1a
|
spectrin repeat containing, nuclear envelope 1a |
| chr15_+_17924404 | 0.23 |
|
|
|
| chr8_-_23591082 | 0.22 |
ENSDART00000025024
|
slc38a5b
|
solute carrier family 38, member 5b |
| chr6_+_32478052 | 0.21 |
|
|
|
| chr17_+_28532738 | 0.20 |
|
|
|
| chr18_+_5616238 | 0.18 |
ENSDART00000163629
|
dut
|
deoxyuridine triphosphatase |
| chr2_+_54075496 | 0.18 |
ENSDART00000166013
|
abhd8a
|
abhydrolase domain containing 8a |
| chr11_-_10867153 | 0.17 |
ENSDART00000091901
|
psmd14
|
proteasome 26S subunit, non-ATPase 14 |
| chr17_+_7437994 | 0.16 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr3_-_20972827 | 0.15 |
ENSDART00000129540
|
larsa
|
leucyl-tRNA synthetase a |
| chr9_-_27939140 | 0.15 |
ENSDART00000064156
|
tbccd1
|
TBCC domain containing 1 |
| chr12_-_31369772 | 0.15 |
ENSDART00000066578
|
tectb
|
tectorin beta |
| chr6_+_41448993 | 0.15 |
ENSDART00000029553
|
rsg1
|
REM2 and RAB-like small GTPase 1 |
| chr19_+_16160028 | 0.12 |
ENSDART00000144376
|
gmeb1
|
glucocorticoid modulatory element binding protein 1 |
| chr24_-_26157275 | 0.10 |
ENSDART00000130696
|
myeov2
|
myeloma overexpressed 2 |
| chr1_+_34823754 | 0.09 |
|
|
|
| chr4_-_16356247 | 0.09 |
ENSDART00000079521
|
kera
|
keratocan |
| chr5_+_42307434 | 0.07 |
|
|
|
| chr3_+_1328162 | 0.06 |
ENSDART00000132817
|
si:ch1073-322p19.1
|
si:ch1073-322p19.1 |
| chr9_-_41072032 | 0.05 |
ENSDART00000066424
|
pofut2
|
protein O-fucosyltransferase 2 |
| chr2_-_11879220 | 0.05 |
ENSDART00000145108
|
ENSDARG00000039203
|
ENSDARG00000039203 |
| chr9_-_35073289 | 0.03 |
ENSDART00000011163
|
asmtl
|
acetylserotonin O-methyltransferase-like |
| chr14_-_6822444 | 0.03 |
ENSDART00000171311
|
si:ch73-43g23.1
|
si:ch73-43g23.1 |
| chr12_-_18839808 | 0.02 |
ENSDART00000172574
|
ep300a
|
E1A binding protein p300 a |
| chr7_-_30353264 | 0.01 |
ENSDART00000173828
|
rnf111
|
ring finger protein 111 |
| chr17_+_7438085 | 0.01 |
ENSDART00000130625
|
si:dkeyp-110a12.4
|
si:dkeyp-110a12.4 |
| chr7_+_17923953 | 0.00 |
ENSDART00000109171
|
rce1a
|
Ras converting CAAX endopeptidase 1a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.4 | 1.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.3 | 2.0 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.3 | 1.3 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.3 | 1.3 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.2 | 0.6 | GO:0010460 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) positive regulation of heart rate(GO:0010460) |
| 0.2 | 0.7 | GO:0061032 | visceral serous pericardium development(GO:0061032) glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.1 | 0.4 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.1 | 0.4 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.1 | 1.4 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 0.8 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) negative regulation of neuron projection regeneration(GO:0070571) |
| 0.1 | 2.8 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 0.8 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.2 | GO:0032329 | serine transport(GO:0032329) L-histidine transmembrane transport(GO:0089709) L-histidine transport(GO:1902024) |
| 0.1 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 2.0 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.1 | 0.4 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.2 | GO:0051645 | Golgi localization(GO:0051645) |
| 0.0 | 0.2 | GO:0098935 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) axo-dendritic protein transport(GO:0099640) |
| 0.0 | 3.0 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 1.3 | GO:1990266 | neutrophil migration(GO:1990266) |
| 0.0 | 0.5 | GO:0001938 | positive regulation of endothelial cell proliferation(GO:0001938) |
| 0.0 | 1.6 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.5 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 1.0 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.5 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 1.2 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.5 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 1.5 | GO:0030239 | myofibril assembly(GO:0030239) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 1.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.0 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 2.8 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.6 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.9 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.6 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 1.0 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 1.0 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.6 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.4 | 1.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.2 | 1.4 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 2.0 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.2 | 1.6 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 0.9 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 1.6 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 0.6 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.4 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 0.3 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.5 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 1.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 0.8 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.0 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.0 | 0.3 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 1.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.2 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.5 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 3.0 | GO:0051015 | actin filament binding(GO:0051015) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.6 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.7 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 1.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.2 | 1.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 1.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 2.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.4 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.3 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.7 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.6 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |