Project
DANIO-CODE
Navigation
Downloads
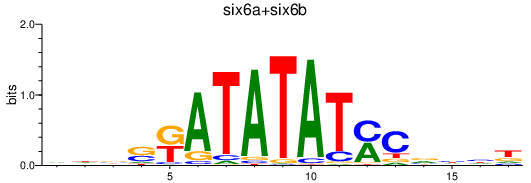
Results for six6a+six6b
Z-value: 0.19
Transcription factors associated with six6a+six6b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
six6a
|
ENSDARG00000025187 | SIX homeobox 6a |
|
six6b
|
ENSDARG00000031316 | SIX homeobox 6b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| six6a | dr10_dc_chr13_+_31473510_31473541 | -0.25 | 3.6e-01 | Click! |
| six6b | dr10_dc_chr20_-_20634087_20634152 | 0.04 | 8.8e-01 | Click! |
Activity profile of six6a+six6b motif
Sorted Z-values of six6a+six6b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of six6a+six6b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_50270783 | 0.18 |
ENSDART00000146056
|
nkx3-1
|
NK3 homeobox 1 |
| chr5_-_11562856 | 0.17 |
ENSDART00000026749
|
nipsnap1
|
nipsnap homolog 1 (C. elegans) |
| chr23_-_45074750 | 0.16 |
ENSDART00000148669
|
si:ch73-269m23.5
|
si:ch73-269m23.5 |
| chr11_-_41357639 | 0.15 |
ENSDART00000055709
|
her2
|
hairy-related 2 |
| chr6_-_16590883 | 0.15 |
ENSDART00000153552
|
nomo
|
nodal modulator |
| chr2_+_10208364 | 0.13 |
|
|
|
| chr5_-_3244261 | 0.11 |
ENSDART00000060162
|
hspb1
|
heat shock protein, alpha-crystallin-related, 1 |
| chr5_-_11562934 | 0.11 |
ENSDART00000026749
|
nipsnap1
|
nipsnap homolog 1 (C. elegans) |
| chr19_+_1298795 | 0.10 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr19_-_18689571 | 0.10 |
ENSDART00000016135
|
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr4_-_13903253 | 0.09 |
ENSDART00000032805
|
gxylt1b
|
glucoside xylosyltransferase 1b |
| chr9_-_306569 | 0.09 |
ENSDART00000166059
|
si:ch211-166e11.5
|
si:ch211-166e11.5 |
| chr23_+_20522512 | 0.09 |
ENSDART00000137294
|
slc35c2
|
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr9_+_50907029 | 0.08 |
|
|
|
| chr2_-_9809756 | 0.08 |
ENSDART00000056899
|
txndc12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr21_+_105327 | 0.08 |
ENSDART00000007672
|
dmgdh
|
dimethylglycine dehydrogenase |
| chr12_+_7220603 | 0.07 |
ENSDART00000092009
|
tfam
|
transcription factor A, mitochondrial |
| chr11_-_17620732 | 0.07 |
ENSDART00000154627
|
eogt
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr20_+_51291139 | 0.07 |
ENSDART00000073981
|
eif2s1b
|
eukaryotic translation initiation factor 2, subunit 1 alpha b |
| chr14_-_21662462 | 0.07 |
ENSDART00000021417
|
p2rx3a
|
purinergic receptor P2X, ligand-gated ion channel, 3a |
| chr19_-_18316262 | 0.07 |
ENSDART00000151133
|
top2b
|
topoisomerase (DNA) II beta |
| chr24_-_14447773 | 0.07 |
|
|
|
| chr22_+_26423320 | 0.07 |
ENSDART00000044085
|
ENSDARG00000034211
|
ENSDARG00000034211 |
| chr8_+_28240085 | 0.07 |
ENSDART00000110857
|
fam212b
|
family with sequence similarity 212, member B |
| chr21_-_3644925 | 0.07 |
ENSDART00000021976
|
nsa2
|
NSA2 ribosome biogenesis homolog (S. cerevisiae) |
| chr24_-_21325777 | 0.07 |
ENSDART00000109848
|
atp8a2
|
ATPase, aminophospholipid transporter, class I, type 8A, member 2 |
| chr24_-_26487093 | 0.07 |
ENSDART00000008374
|
tnikb
|
TRAF2 and NCK interacting kinase b |
| chr6_-_9410600 | 0.07 |
ENSDART00000012903
|
wdr12
|
WD repeat domain 12 |
| chr14_+_26461399 | 0.06 |
ENSDART00000125289
|
ahnak
|
AHNAK nucleoprotein |
| chr10_-_5820321 | 0.06 |
ENSDART00000172409
|
il6st
|
interleukin 6 signal transducer |
| chr9_+_907908 | 0.06 |
ENSDART00000144114
|
dbi
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr15_-_25592228 | 0.06 |
ENSDART00000157498
|
hif1al
|
hypoxia-inducible factor 1, alpha subunit, like |
| chr8_-_8307383 | 0.05 |
|
|
|
| chr10_+_36751832 | 0.05 |
ENSDART00000169015
|
rab6a
|
RAB6A, member RAS oncogene family |
| chr4_-_25869361 | 0.05 |
ENSDART00000128968
|
nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr3_+_23608395 | 0.05 |
|
|
|
| chr19_+_41894906 | 0.05 |
ENSDART00000087187
|
ago2
|
argonaute RISC catalytic component 2 |
| chr23_-_7892862 | 0.05 |
ENSDART00000157612
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr18_+_20045221 | 0.05 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr8_+_2517472 | 0.05 |
ENSDART00000112914
|
si:ch211-51h9.7
|
si:ch211-51h9.7 |
| chr17_-_23235070 | 0.05 |
|
|
|
| chr15_-_784846 | 0.05 |
ENSDART00000155533
|
im:7143333
|
im:7143333 |
| chr13_+_44720106 | 0.05 |
ENSDART00000017770
|
zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr7_+_67510360 | 0.05 |
ENSDART00000172015
|
cyb5b
|
cytochrome b5 type B |
| chr16_-_4940567 | 0.05 |
ENSDART00000149421
|
rpa2
|
replication protein A2 |
| chr23_+_32409339 | 0.04 |
ENSDART00000149698
|
slc39a5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr22_+_19615230 | 0.04 |
ENSDART00000061725
ENSDART00000140819 |
rgmd
|
RGM domain family, member D |
| chr20_-_9593191 | 0.04 |
ENSDART00000014168
|
zfp36l1b
|
zinc finger protein 36, C3H type-like 1b |
| chr8_+_28377022 | 0.04 |
ENSDART00000158788
|
klhl12
|
kelch-like family member 12 |
| chr24_+_2322328 | 0.04 |
ENSDART00000132530
|
si:ch211-266d19.4
|
si:ch211-266d19.4 |
| chr18_-_14709371 | 0.04 |
ENSDART00000111995
|
si:dkey-238o13.4
|
si:dkey-238o13.4 |
| chr25_+_558191 | 0.04 |
ENSDART00000126863
ENSDART00000166012 |
nell2a
|
neural EGFL like 2a |
| chr9_-_11705194 | 0.04 |
ENSDART00000022358
|
zc3h15
|
zinc finger CCCH-type containing 15 |
| chr14_+_26461172 | 0.04 |
ENSDART00000125289
|
ahnak
|
AHNAK nucleoprotein |
| chr4_-_71612399 | 0.04 |
ENSDART00000158194
|
wu:fc30c06
|
wu:fc30c06 |
| chr17_-_610651 | 0.04 |
|
|
|
| chr1_-_34921848 | 0.04 |
ENSDART00000142154
|
frem3
|
Fras1 related extracellular matrix 3 |
| chr3_-_19968543 | 0.04 |
ENSDART00000055745
ENSDART00000139902 |
sepw2a
|
selenoprotein W, 2a |
| chr2_-_30266151 | 0.04 |
ENSDART00000142575
|
tceb1b
|
transcription elongation factor B (SIII), polypeptide 1b |
| chr6_-_48419131 | 0.04 |
ENSDART00000090528
|
rhoca
|
ras homolog family member Ca |
| chr4_+_1911689 | 0.03 |
ENSDART00000026963
|
pmpcb
|
peptidase (mitochondrial processing) beta |
| chr16_-_37025649 | 0.03 |
ENSDART00000113834
|
snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12) |
| KN150196v1_+_15525 | 0.03 |
|
|
|
| chr16_+_54001592 | 0.03 |
|
|
|
| chr10_+_10428297 | 0.03 |
ENSDART00000179214
|
sardh
|
sarcosine dehydrogenase |
| chr10_-_3260827 | 0.03 |
ENSDART00000113162
|
pi4kaa
|
phosphatidylinositol 4-kinase, catalytic, alpha a |
| chr25_-_10533634 | 0.03 |
ENSDART00000153639
|
ppp6r3
|
protein phosphatase 6, regulatory subunit 3 |
| chr1_-_11474337 | 0.03 |
ENSDART00000149913
|
b4galt1
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 |
| chr3_+_37565550 | 0.03 |
ENSDART00000151236
|
map3k14a
|
mitogen-activated protein kinase kinase kinase 14a |
| chr22_+_26543146 | 0.03 |
|
|
|
| KN150071v1_+_374 | 0.03 |
ENSDART00000172649
|
CABZ01033178.1
|
ENSDARG00000103512 |
| chr25_+_3180913 | 0.03 |
ENSDART00000034704
|
slc25a3b
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3b |
| chr13_-_48884956 | 0.03 |
|
|
|
| chr25_-_34542816 | 0.03 |
ENSDART00000155060
|
zgc:110434
|
zgc:110434 |
| chr13_+_22425272 | 0.03 |
ENSDART00000133617
|
bmpr1aa
|
bone morphogenetic protein receptor, type IAa |
| chr13_+_35619748 | 0.03 |
ENSDART00000159690
|
gpr75
|
G protein-coupled receptor 75 |
| chr21_-_1618494 | 0.03 |
ENSDART00000124904
|
zgc:152948
|
zgc:152948 |
| chr1_-_44922521 | 0.02 |
ENSDART00000149725
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr1_-_30925256 | 0.02 |
ENSDART00000085309
|
dpcd
|
deleted in primary ciliary dyskinesia homolog (mouse) |
| chr15_-_35075545 | 0.02 |
ENSDART00000006288
|
dhx16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr2_+_12547126 | 0.02 |
ENSDART00000169724
|
zgc:66475
|
zgc:66475 |
| chr18_+_12089651 | 0.02 |
ENSDART00000140854
|
bicd1a
|
bicaudal D homolog 1a |
| chr20_+_37480145 | 0.02 |
ENSDART00000134997
ENSDART00000145927 |
adgrg6
|
adhesion G protein-coupled receptor G6 |
| chr25_-_19476079 | 0.02 |
ENSDART00000156811
|
si:ch211-59o9.10
|
si:ch211-59o9.10 |
| chr12_+_34631742 | 0.02 |
|
|
|
| chr2_+_57903155 | 0.02 |
|
|
|
| chr2_-_55565099 | 0.02 |
ENSDART00000149062
|
rab8a
|
RAB8A, member RAS oncogene family |
| chr1_+_52739724 | 0.02 |
ENSDART00000159220
|
ucp1
|
uncoupling protein 1 |
| chr20_-_34808532 | 0.02 |
ENSDART00000061555
|
si:ch211-63o20.7
|
si:ch211-63o20.7 |
| chr24_-_31285903 | 0.02 |
ENSDART00000164001
|
alg14
|
ALG14, UDP-N-acetylglucosaminyltransferase subunit |
| KN150127v1_-_32012 | 0.02 |
|
|
|
| chr12_-_926641 | 0.02 |
ENSDART00000088351
|
SPAG9 (1 of many)
|
sperm associated antigen 9 |
| chr9_-_24407859 | 0.02 |
ENSDART00000135356
|
nabp1a
|
nucleic acid binding protein 1a |
| chr17_+_28689850 | 0.02 |
ENSDART00000126967
|
strn3
|
striatin, calmodulin binding protein 3 |
| chr24_-_39884167 | 0.02 |
ENSDART00000087441
|
GFOD1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr24_+_16861210 | 0.02 |
ENSDART00000149149
|
zfx
|
zinc finger protein, X-linked |
| chr21_-_30977021 | 0.02 |
ENSDART00000065504
|
ncbp3
|
nuclear cap binding subunit 3 |
| chr22_-_2912805 | 0.02 |
ENSDART00000178290
|
pak2b
|
p21 protein (Cdc42/Rac)-activated kinase 2b |
| chr18_+_14725148 | 0.02 |
ENSDART00000146128
|
uri1
|
URI1, prefoldin-like chaperone |
| chr2_+_30266562 | 0.02 |
ENSDART00000135171
|
tmem70
|
transmembrane protein 70 |
| chr24_-_25643778 | 0.02 |
ENSDART00000015628
ENSDART00000132235 |
klhl24b
|
kelch-like family member 24b |
| chr19_+_41894734 | 0.02 |
ENSDART00000087187
|
ago2
|
argonaute RISC catalytic component 2 |
| chr18_-_17735227 | 0.02 |
|
|
|
| chr10_+_45056951 | 0.02 |
ENSDART00000158553
|
ENSDARG00000101143
|
ENSDARG00000101143 |
| chr1_-_55211232 | 0.02 |
ENSDART00000064194
|
asf1bb
|
anti-silencing function 1Bb histone chaperone |
| chr24_-_14447655 | 0.01 |
|
|
|
| chr9_+_54736764 | 0.01 |
ENSDART00000148928
|
rbm27
|
RNA binding motif protein 27 |
| chr17_+_37306052 | 0.01 |
ENSDART00000134000
|
tmem62
|
transmembrane protein 62 |
| chr13_+_3750263 | 0.01 |
ENSDART00000169595
|
timm23b
|
translocase of inner mitochondrial membrane 23 homolog b (yeast) |
| chr12_+_10668405 | 0.01 |
ENSDART00000161986
ENSDART00000158227 |
top2a
|
topoisomerase (DNA) II alpha |
| chr2_+_37262863 | 0.01 |
ENSDART00000143311
|
sec62
|
SEC62 homolog, preprotein translocation factor |
| chr4_-_4242819 | 0.01 |
|
|
|
| chr10_-_35377693 | 0.01 |
ENSDART00000131268
|
rhbdd2
|
rhomboid domain containing 2 |
| chr5_-_29916352 | 0.01 |
ENSDART00000014666
|
arcn1a
|
archain 1a |
| chr7_-_7533548 | 0.01 |
ENSDART00000173376
|
intu
|
inturned planar cell polarity protein |
| chr15_-_35020312 | 0.01 |
ENSDART00000154094
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr21_-_26214047 | 0.01 |
ENSDART00000044735
|
gria1b
|
glutamate receptor, ionotropic, AMPA 1b |
| chr19_-_1921261 | 0.01 |
ENSDART00000004585
|
clptm1l
|
CLPTM1-like |
| chr15_-_723851 | 0.01 |
|
|
|
| chr19_-_33123907 | 0.01 |
ENSDART00000004034
|
hpca
|
hippocalcin |
| chr9_+_28420988 | 0.01 |
|
|
|
| chr18_+_14725283 | 0.01 |
ENSDART00000146128
|
uri1
|
URI1, prefoldin-like chaperone |
| chr18_-_25784699 | 0.01 |
ENSDART00000103046
|
zgc:162879
|
zgc:162879 |
| chr1_+_30925269 | 0.01 |
ENSDART00000057880
|
poll
|
polymerase (DNA directed), lambda |
| chr15_-_25158683 | 0.01 |
ENSDART00000129154
|
exo5
|
exonuclease 5 |
| chr13_-_34666736 | 0.01 |
|
|
|
| chr13_-_24130160 | 0.01 |
ENSDART00000138747
ENSDART00000101168 |
urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr4_+_23396557 | 0.01 |
ENSDART00000066909
|
slc35e3
|
solute carrier family 35, member E3 |
| chr22_+_2231377 | 0.01 |
ENSDART00000143366
|
si:dkey-4c15.8
|
si:dkey-4c15.8 |
| chr15_-_35020250 | 0.01 |
ENSDART00000154094
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr25_+_34650598 | 0.01 |
ENSDART00000114598
|
ano9a
|
anoctamin 9a |
| chr22_-_12837665 | 0.01 |
ENSDART00000145156
ENSDART00000137280 |
glsa
|
glutaminase a |
| chr6_-_43782889 | 0.01 |
|
|
|
| chr7_-_6289729 | 0.00 |
ENSDART00000173323
|
hist1h4l
|
histone 1, H4, like |
| chr7_-_69883581 | 0.00 |
|
|
|
| chr18_+_16144493 | 0.00 |
ENSDART00000080423
|
ctsd
|
cathepsin D |
| chr14_-_17270741 | 0.00 |
ENSDART00000161355
ENSDART00000168959 |
rnf4
|
ring finger protein 4 |
| KN150438v1_+_427 | 0.00 |
|
|
|
| chr19_+_390050 | 0.00 |
ENSDART00000178634
|
sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr18_+_44680485 | 0.00 |
ENSDART00000018625
|
napab
|
N-ethylmaleimide-sensitive factor attachment protein, alpha b |
| chr2_-_45093067 | 0.00 |
ENSDART00000036997
|
camk2n1a
|
calcium/calmodulin-dependent protein kinase II inhibitor 1a |
| chr21_-_9469489 | 0.00 |
ENSDART00000162225
ENSDART00000163874 ENSDART00000168173 |
ptpn13
|
protein tyrosine phosphatase, non-receptor type 13 |
| chr20_-_5355373 | 0.00 |
ENSDART00000168103
|
snw1
|
SNW domain containing 1 |
| chr1_+_43347121 | 0.00 |
|
|
|
| chr6_-_50686517 | 0.00 |
ENSDART00000134146
|
mtss1
|
metastasis suppressor 1 |
| chr10_+_38764480 | 0.00 |
ENSDART00000172306
ENSDART00000163099 |
tmprss2
|
transmembrane protease, serine 2 |
| chr7_+_46964914 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0038107 | nodal signaling pathway involved in determination of left/right asymmetry(GO:0038107) regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) |
| 0.0 | 0.1 | GO:0061687 | detoxification of inorganic compound(GO:0061687) |
| 0.0 | 0.2 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0014881 | regulation of myofibril size(GO:0014881) response to arsenic-containing substance(GO:0046685) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.0 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.0 | 0.1 | GO:0033198 | response to ATP(GO:0033198) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0046997 | oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.0 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.1 | GO:0016502 | purinergic nucleotide receptor activity(GO:0001614) extracellular ATP-gated cation channel activity(GO:0004931) nucleotide receptor activity(GO:0016502) ATP-gated ion channel activity(GO:0035381) |