Project
DANIO-CODE
Navigation
Downloads
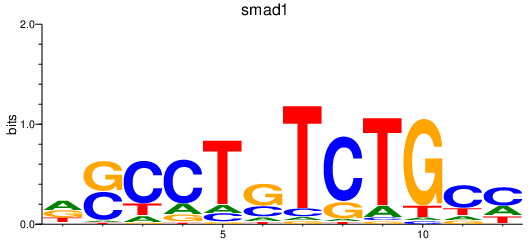
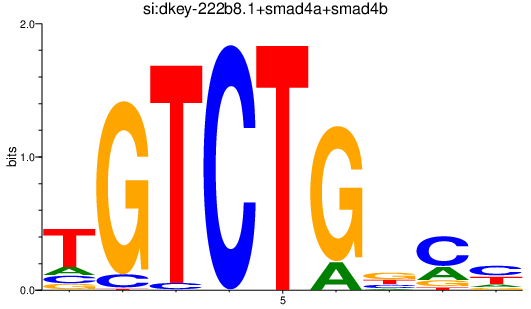
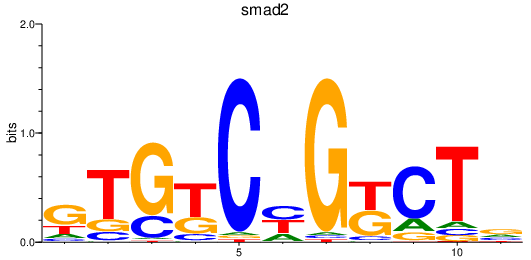
Results for smad1_si:dkey-222b8.1+smad4a+smad4b_smad2
Z-value: 1.02
Transcription factors associated with smad1_si:dkey-222b8.1+smad4a+smad4b_smad2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
smad1
|
ENSDARG00000027199 | SMAD family member 1 |
|
smad4b
|
ENSDARG00000012649 | SMAD family member 4b |
|
si_dkey-222b8.1
|
ENSDARG00000045094 | si_dkey-222b8.1 |
|
smad4a
|
ENSDARG00000075226 | SMAD family member 4a |
|
smad2
|
ENSDARG00000006389 | SMAD family member 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| smad2 | dr10_dc_chr10_-_14971565_14971593 | -0.83 | 7.7e-05 | Click! |
| smad1 | dr10_dc_chr1_-_35196892_35196970 | 0.77 | 5.3e-04 | Click! |
| SMAD4 (1 of many) | dr10_dc_chr5_+_6308337_6308415 | 0.71 | 2.0e-03 | Click! |
| smad4a | dr10_dc_chr10_+_573116_573138 | -0.60 | 1.4e-02 | Click! |
Activity profile of smad1_si:dkey-222b8.1+smad4a+smad4b_smad2 motif
Sorted Z-values of smad1_si:dkey-222b8.1+smad4a+smad4b_smad2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of smad1_si:dkey-222b8.1+smad4a+smad4b_smad2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_52872108 | 6.02 |
ENSDART00000157453
ENSDART00000163684 |
nme8
|
NME/NM23 family member 8 |
| chr23_-_39956151 | 3.61 |
ENSDART00000115330
|
ppp1r14c
|
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr5_-_65349550 | 2.86 |
ENSDART00000164228
|
nrarpb
|
notch-regulated ankyrin repeat protein b |
| chr20_+_18263793 | 2.69 |
ENSDART00000011287
|
aqp4
|
aquaporin 4 |
| chr18_-_44914637 | 2.31 |
ENSDART00000169636
|
si:ch211-71n6.4
|
si:ch211-71n6.4 |
| chr15_+_40161311 | 2.28 |
ENSDART00000063783
|
itm2ca
|
integral membrane protein 2Ca |
| chr6_+_7936632 | 2.17 |
ENSDART00000146106
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr16_-_38117896 | 2.02 |
ENSDART00000114266
|
plekho1b
|
pleckstrin homology domain containing, family O member 1b |
| chr17_-_21260319 | 1.87 |
ENSDART00000166524
|
hspa12a
|
heat shock protein 12A |
| chr12_-_9256949 | 1.85 |
ENSDART00000003805
|
pth1rb
|
parathyroid hormone 1 receptor b |
| chr4_-_4923982 | 1.70 |
ENSDART00000103293
|
ndufa5
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5 |
| chr2_-_52202004 | 1.69 |
ENSDART00000165350
|
BX908782.1
|
ENSDARG00000098957 |
| chr10_-_10059864 | 1.68 |
ENSDART00000132375
|
strbp
|
spermatid perinuclear RNA binding protein |
| chr24_+_40625215 | 1.67 |
|
|
|
| chr10_+_20406765 | 1.60 |
ENSDART00000080395
|
golga7
|
golgin A7 |
| KN150142v1_-_9787 | 1.56 |
ENSDART00000166321
ENSDART00000170296 |
ENSDARG00000100534
|
ENSDARG00000100534 |
| chr13_-_349685 | 1.46 |
ENSDART00000159803
|
heatr5b
|
HEAT repeat containing 5B |
| chr18_+_49230636 | 1.44 |
ENSDART00000167609
ENSDART00000135026 ENSDART00000171618 |
si:ch211-136a13.1
|
si:ch211-136a13.1 |
| chr1_+_25717717 | 1.40 |
ENSDART00000112263
|
arhgef38
|
Rho guanine nucleotide exchange factor (GEF) 38 |
| chr19_+_26838851 | 1.39 |
ENSDART00000136244
|
npr1a
|
natriuretic peptide receptor 1a |
| chr12_-_17590507 | 1.36 |
ENSDART00000010144
|
pvalb2
|
parvalbumin 2 |
| chr25_+_31547276 | 1.33 |
ENSDART00000090727
|
duox
|
dual oxidase |
| chr19_-_41482388 | 1.32 |
ENSDART00000111982
|
sgce
|
sarcoglycan, epsilon |
| chr20_-_16022816 | 1.31 |
ENSDART00000152828
|
fam20b
|
family with sequence similarity 20, member B (H. sapiens) |
| chr21_-_7703717 | 1.27 |
ENSDART00000158852
|
egfl7
|
EGF-like-domain, multiple 7 |
| chr23_+_29981525 | 1.23 |
ENSDART00000035755
|
dvl1a
|
dishevelled segment polarity protein 1a |
| chr8_+_7882144 | 1.21 |
|
|
|
| chr19_-_33123724 | 1.20 |
ENSDART00000004034
|
hpca
|
hippocalcin |
| chr18_+_618393 | 1.18 |
ENSDART00000159464
|
nedd4a
|
neural precursor cell expressed, developmentally down-regulated 4a |
| chr25_+_222244 | 1.17 |
ENSDART00000155344
|
ENSDARG00000073905
|
ENSDARG00000073905 |
| chr25_+_7000082 | 1.16 |
|
|
|
| chr12_-_5470187 | 1.12 |
ENSDART00000159784
|
abi3b
|
ABI family, member 3b |
| chr21_+_17073163 | 1.10 |
|
|
|
| chr15_+_28370102 | 1.05 |
ENSDART00000175860
|
myo1cb
|
myosin Ic, paralog b |
| chr23_+_41015697 | 1.03 |
ENSDART00000115161
|
reps2
|
RALBP1 associated Eps domain containing 2 |
| chr6_+_7936373 | 1.03 |
ENSDART00000146106
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr5_+_6054781 | 1.03 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr20_-_31524559 | 1.01 |
ENSDART00000007735
|
ust
|
uronyl-2-sulfotransferase |
| chr14_+_6239755 | 1.00 |
ENSDART00000097214
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr3_+_32394653 | 0.98 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr5_+_34397578 | 0.98 |
ENSDART00000043341
|
foxd1
|
forkhead box D1 |
| chr22_+_38837247 | 0.97 |
ENSDART00000144318
|
anos1b
|
anosmin 1b |
| chr25_-_3766929 | 0.97 |
ENSDART00000043172
|
tmem258
|
transmembrane protein 258 |
| chr10_+_25304728 | 0.97 |
ENSDART00000100438
|
rab38b
|
RAB38b, member of RAS oncogene family |
| chr5_-_37850885 | 0.95 |
ENSDART00000136428
ENSDART00000112487 |
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr22_-_520129 | 0.93 |
ENSDART00000001201
|
bysl
|
bystin-like |
| chr4_+_22959458 | 0.90 |
ENSDART00000036531
|
gnai1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr21_-_3301751 | 0.89 |
ENSDART00000009740
|
smad7
|
SMAD family member 7 |
| chr2_+_33474083 | 0.84 |
ENSDART00000056657
|
zgc:113531
|
zgc:113531 |
| chr23_+_157661 | 0.84 |
|
|
|
| chr23_+_36832325 | 0.83 |
|
|
|
| chr3_+_25888520 | 0.83 |
ENSDART00000135389
|
foxred2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr25_+_31746643 | 0.83 |
ENSDART00000162636
|
tjp1b
|
tight junction protein 1b |
| chr19_-_44223619 | 0.82 |
ENSDART00000160879
|
klhl43
|
kelch-like family member 43 |
| chr8_+_26273354 | 0.82 |
ENSDART00000053463
|
mgll
|
monoglyceride lipase |
| chr8_-_17480730 | 0.82 |
ENSDART00000100667
|
skia
|
v-ski avian sarcoma viral oncogene homolog a |
| chr11_+_39768718 | 0.78 |
ENSDART00000130278
|
si:dkey-264d12.1
|
si:dkey-264d12.1 |
| chr24_+_40974634 | 0.77 |
|
|
|
| chr5_-_37851313 | 0.77 |
ENSDART00000136428
ENSDART00000112487 |
chrne
|
cholinergic receptor, nicotinic, epsilon |
| chr12_-_4033069 | 0.77 |
ENSDART00000042200
|
aldoab
|
aldolase a, fructose-bisphosphate, b |
| chr25_-_30818836 | 0.75 |
ENSDART00000156828
|
prr33
|
proline rich 33 |
| chr10_-_43506501 | 0.75 |
ENSDART00000146660
|
hapln1b
|
hyaluronan and proteoglycan link protein 1b |
| chr14_-_26999410 | 0.74 |
ENSDART00000159727
|
pcdh11
|
protocadherin 11 |
| chr19_-_5464238 | 0.74 |
ENSDART00000105806
|
cyt1
|
type I cytokeratin, enveloping layer |
| chr4_-_25869361 | 0.74 |
ENSDART00000128968
|
nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr21_+_45227200 | 0.73 |
ENSDART00000155681
ENSDART00000157136 |
tcf7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr3_+_16115532 | 0.73 |
|
|
|
| chr8_-_12365342 | 0.72 |
ENSDART00000113286
|
phf19
|
PHD finger protein 19 |
| chr19_-_2939818 | 0.72 |
|
|
|
| chr23_-_27226280 | 0.72 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr9_+_3147601 | 0.71 |
ENSDART00000135619
|
ftr52p
|
finTRIM family, member 52, pseudogene |
| chr19_-_47897712 | 0.70 |
|
|
|
| chr18_+_23004984 | 0.70 |
ENSDART00000171871
|
cbfb
|
core-binding factor, beta subunit |
| chr10_-_20489955 | 0.68 |
ENSDART00000159672
|
loxl2a
|
lysyl oxidase-like 2a |
| chr1_+_1657493 | 0.66 |
ENSDART00000103850
|
atp1a1a.3
|
ATPase, Na+/K+ transporting, alpha 1a polypeptide, tandem duplicate 3 |
| chr4_-_26333625 | 0.66 |
ENSDART00000170709
|
st8sia1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr25_+_30535271 | 0.66 |
|
|
|
| chr20_-_31524614 | 0.65 |
ENSDART00000007735
|
ust
|
uronyl-2-sulfotransferase |
| chr14_+_9115745 | 0.65 |
ENSDART00000123652
|
hmgn6
|
high mobility group nucleosome binding domain 6 |
| chr13_+_48324283 | 0.65 |
ENSDART00000177067
|
CU929120.1
|
ENSDARG00000107321 |
| chr9_+_42112576 | 0.64 |
ENSDART00000130434
ENSDART00000007058 |
col18a1a
|
collagen type XVIII alpha 1 chain a |
| chr24_+_38371710 | 0.63 |
ENSDART00000105672
|
mybpc2b
|
myosin binding protein C, fast type b |
| chr13_+_45843567 | 0.62 |
ENSDART00000005195
ENSDART00000074547 |
bsdc1
|
BSD domain containing 1 |
| chr2_-_59531831 | 0.61 |
ENSDART00000168568
|
CU861477.1
|
ENSDARG00000100128 |
| chr16_-_8749368 | 0.61 |
ENSDART00000162988
|
cobl
|
cordon-bleu WH2 repeat protein |
| chr21_+_5887486 | 0.61 |
ENSDART00000048399
|
slc4a4b
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4b |
| chr4_+_10065500 | 0.61 |
ENSDART00000026492
|
flncb
|
filamin C, gamma b (actin binding protein 280) |
| chr22_-_11106940 | 0.61 |
ENSDART00000016873
|
atp6ap2
|
ATPase, H+ transporting, lysosomal accessory protein 2 |
| chr13_+_23065500 | 0.61 |
ENSDART00000158370
|
sorbs1
|
sorbin and SH3 domain containing 1 |
| chr20_+_34497817 | 0.60 |
ENSDART00000061632
|
fam129aa
|
family with sequence similarity 129, member Aa |
| chr16_+_24045774 | 0.58 |
ENSDART00000133484
|
apoeb
|
apolipoprotein Eb |
| chr23_+_2101659 | 0.58 |
|
|
|
| chr5_-_71340996 | 0.58 |
ENSDART00000162526
|
smyd1a
|
SET and MYND domain containing 1a |
| chr15_+_34131126 | 0.57 |
ENSDART00000169487
|
vwde
|
von Willebrand factor D and EGF domains |
| chr8_+_28493742 | 0.57 |
ENSDART00000053782
|
scrt2
|
scratch family zinc finger 2 |
| KN150670v1_-_72156 | 0.56 |
|
|
|
| chr11_-_23151247 | 0.56 |
|
|
|
| chr19_-_325337 | 0.56 |
ENSDART00000147635
|
gpd1c
|
glycerol-3-phosphate dehydrogenase 1c |
| chr7_-_72127164 | 0.56 |
ENSDART00000176069
|
CABZ01057364.1
|
ENSDARG00000106736 |
| chr10_+_4874841 | 0.54 |
ENSDART00000165942
|
palm2
|
paralemmin 2 |
| chr7_-_25623974 | 0.54 |
ENSDART00000173602
|
cd99l2
|
CD99 molecule-like 2 |
| chr2_-_7693224 | 0.54 |
ENSDART00000167019
|
wu:fc46h12
|
wu:fc46h12 |
| chr8_-_38357549 | 0.54 |
ENSDART00000129597
|
sorbs3
|
sorbin and SH3 domain containing 3 |
| chr9_-_33584190 | 0.53 |
ENSDART00000138844
|
caska
|
calcium/calmodulin-dependent serine protein kinase a |
| chr2_+_49358871 | 0.53 |
ENSDART00000179089
|
sema6ba
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6Ba |
| chr3_+_24067387 | 0.53 |
ENSDART00000055609
|
atf4b
|
activating transcription factor 4b |
| chr21_-_26459113 | 0.52 |
ENSDART00000157255
|
cd248b
|
CD248 molecule, endosialin b |
| chr4_-_18606513 | 0.52 |
ENSDART00000049061
|
cdkn1ba
|
cyclin-dependent kinase inhibitor 1Ba |
| chr21_+_43674754 | 0.52 |
ENSDART00000136025
|
tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr23_-_5429419 | 0.52 |
ENSDART00000159105
|
CR936540.3
|
ENSDARG00000101435 |
| chr12_+_23717554 | 0.51 |
ENSDART00000152997
|
svila
|
supervillin a |
| chr19_-_33123907 | 0.51 |
ENSDART00000004034
|
hpca
|
hippocalcin |
| chr11_-_2107054 | 0.51 |
ENSDART00000173031
|
hoxc6b
|
homeobox C6b |
| chr23_-_27226387 | 0.50 |
ENSDART00000141305
|
si:dkey-157g16.6
|
si:dkey-157g16.6 |
| chr3_-_53304694 | 0.50 |
ENSDART00000073930
|
notch3
|
notch 3 |
| chr10_+_33926312 | 0.49 |
ENSDART00000174730
|
BX569785.2
|
ENSDARG00000106629 |
| chr19_+_41966252 | 0.49 |
ENSDART00000169193
|
si:ch211-57n23.4
|
si:ch211-57n23.4 |
| chr17_-_14663139 | 0.49 |
ENSDART00000037371
|
ppp1r13ba
|
protein phosphatase 1, regulatory subunit 13Ba |
| chr14_-_46648563 | 0.49 |
|
|
|
| chr16_-_26258657 | 0.48 |
ENSDART00000157787
|
lipeb
|
lipase, hormone-sensitive b |
| chr17_+_33481064 | 0.48 |
ENSDART00000077581
|
snap23.2
|
synaptosomal-associated protein 23.2 |
| chr6_-_4369754 | 0.48 |
|
|
|
| chr6_+_7936576 | 0.48 |
ENSDART00000146106
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr25_-_30856032 | 0.48 |
ENSDART00000067028
|
zgc:172145
|
zgc:172145 |
| chr4_-_74907755 | 0.47 |
ENSDART00000132323
|
ftr51
|
finTRIM family, member 51 |
| chr7_+_40183670 | 0.47 |
ENSDART00000173916
|
ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2 |
| chr2_-_31037043 | 0.47 |
ENSDART00000087026
|
emilin2a
|
elastin microfibril interfacer 2a |
| chr1_-_50513645 | 0.47 |
ENSDART00000047851
|
jag1a
|
jagged 1a |
| chr10_-_44117211 | 0.46 |
ENSDART00000009134
|
sept5b
|
septin 5b |
| chr13_-_49826330 | 0.46 |
ENSDART00000099475
|
nid1a
|
nidogen 1a |
| chr6_+_6333766 | 0.46 |
ENSDART00000140827
|
bcl11ab
|
B-cell CLL/lymphoma 11Ab |
| chr8_+_47644421 | 0.45 |
ENSDART00000139096
|
si:ch211-251b21.1
|
si:ch211-251b21.1 |
| chr24_-_36287207 | 0.45 |
ENSDART00000065338
|
pak1ip1
|
PAK1 interacting protein 1 |
| chr3_-_36704626 | 0.45 |
ENSDART00000055237
|
ramp2
|
receptor (G protein-coupled) activity modifying protein 2 |
| chr6_-_9964616 | 0.45 |
|
|
|
| chr6_-_39311711 | 0.45 |
|
|
|
| chr10_+_18994733 | 0.45 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr12_+_30538113 | 0.45 |
ENSDART00000179355
|
thbs2b
|
thrombospondin 2b |
| chr19_+_2162466 | 0.45 |
|
|
|
| chr23_-_15442032 | 0.45 |
ENSDART00000082060
|
ENSDARG00000078145
|
ENSDARG00000078145 |
| chr3_+_14238188 | 0.45 |
ENSDART00000165452
ENSDART00000171726 |
tmem56b
|
transmembrane protein 56b |
| chr14_-_9114872 | 0.44 |
ENSDART00000081158
|
sh3bgrl
|
SH3 domain binding glutamate-rich protein like |
| chr5_+_34397618 | 0.44 |
ENSDART00000043341
|
foxd1
|
forkhead box D1 |
| chr9_-_21420106 | 0.44 |
ENSDART00000102147
|
popdc2
|
popeye domain containing 2 |
| chr13_+_45843396 | 0.43 |
ENSDART00000005195
ENSDART00000074547 |
bsdc1
|
BSD domain containing 1 |
| chr14_-_12001505 | 0.43 |
ENSDART00000171136
|
myot
|
myotilin |
| chr9_+_51693805 | 0.43 |
ENSDART00000132896
|
fap
|
fibroblast activation protein, alpha |
| chr21_-_11539798 | 0.43 |
ENSDART00000144770
|
cast
|
calpastatin |
| chr23_+_36007936 | 0.42 |
ENSDART00000128533
|
hoxc3a
|
homeo box C3a |
| chr24_-_41448993 | 0.42 |
ENSDART00000041349
|
crygn2
|
crystallin, gamma N2 |
| chr1_-_29669484 | 0.42 |
ENSDART00000164202
|
igf2bp2b
|
insulin-like growth factor 2 mRNA binding protein 2b |
| chr6_+_41466199 | 0.42 |
ENSDART00000177439
|
twf2a
|
twinfilin actin-binding protein 2a |
| chr18_-_44135616 | 0.42 |
ENSDART00000087339
|
cdon
|
cell adhesion associated, oncogene regulated |
| chr17_+_22082952 | 0.42 |
ENSDART00000047772
|
mal
|
mal, T-cell differentiation protein |
| chr9_+_308260 | 0.41 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr5_-_34397435 | 0.41 |
ENSDART00000170684
|
btf3
|
basic transcription factor 3 |
| chr20_+_35010140 | 0.41 |
ENSDART00000172001
|
snap25a
|
synaptosomal-associated protein, 25a |
| chr7_+_23778570 | 0.41 |
ENSDART00000121837
|
efs
|
embryonal Fyn-associated substrate |
| chr23_-_42775849 | 0.41 |
ENSDART00000149944
|
gpx7
|
glutathione peroxidase 7 |
| chr23_-_34970838 | 0.41 |
ENSDART00000087219
|
ENSDARG00000061272
|
ENSDARG00000061272 |
| chr13_-_25067585 | 0.41 |
ENSDART00000159585
|
adka
|
adenosine kinase a |
| chr6_+_47845002 | 0.41 |
ENSDART00000140943
|
padi2
|
peptidyl arginine deiminase, type II |
| chr23_+_45020761 | 0.41 |
ENSDART00000159104
|
atp1b2a
|
ATPase, Na+/K+ transporting, beta 2a polypeptide |
| chr15_-_8333117 | 0.40 |
ENSDART00000061351
|
tnfrsf19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr3_+_34064081 | 0.40 |
|
|
|
| chr2_-_59047356 | 0.40 |
|
|
|
| chr21_+_44749057 | 0.40 |
ENSDART00000084780
|
rnf121
|
ring finger protein 121 |
| chr5_+_27298136 | 0.40 |
ENSDART00000098604
|
adam28
|
ADAM metallopeptidase domain 28 |
| chr25_-_6501787 | 0.39 |
ENSDART00000112782
|
cspg4
|
chondroitin sulfate proteoglycan 4 |
| chr6_-_8126955 | 0.39 |
ENSDART00000132412
|
acp5a
|
acid phosphatase 5a, tartrate resistant |
| chr4_+_57435 | 0.39 |
ENSDART00000169187
|
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr7_+_34417030 | 0.39 |
ENSDART00000108473
|
plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr9_+_54692834 | 0.39 |
|
|
|
| chr8_-_19364689 | 0.39 |
ENSDART00000014183
|
colgalt2
|
collagen beta(1-O)galactosyltransferase 2 |
| KN150495v1_-_20497 | 0.39 |
|
|
|
| chr1_-_50513873 | 0.39 |
ENSDART00000137172
|
jag1a
|
jagged 1a |
| chr1_-_50066633 | 0.39 |
|
|
|
| chr21_+_28459490 | 0.39 |
ENSDART00000138236
|
cox8a
|
cytochrome c oxidase subunit VIIIA (ubiquitous) |
| chr8_-_40231417 | 0.38 |
ENSDART00000162020
|
kdm2ba
|
lysine (K)-specific demethylase 2Ba |
| chr25_-_32344881 | 0.38 |
ENSDART00000012862
|
isl2a
|
ISL LIM homeobox 2a |
| chr20_-_7303804 | 0.38 |
ENSDART00000100060
|
dsc2l
|
desmocollin 2 like |
| chr5_-_25866099 | 0.38 |
ENSDART00000144035
|
arvcfb
|
armadillo repeat gene deleted in velocardiofacial syndrome b |
| chr9_+_24089828 | 0.38 |
ENSDART00000127859
|
trim63b
|
tripartite motif containing 63b |
| chr6_-_39315024 | 0.38 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr2_+_55888093 | 0.38 |
ENSDART00000146160
|
loxl5b
|
lysyl oxidase-like 5b |
| chr3_-_39313029 | 0.38 |
ENSDART00000156123
|
CU464118.1
|
ENSDARG00000097747 |
| chr19_+_12663944 | 0.37 |
ENSDART00000151508
|
ldlrad4a
|
low density lipoprotein receptor class A domain containing 4a |
| chr16_+_74373 | 0.37 |
|
|
|
| chr15_+_32963784 | 0.37 |
ENSDART00000167739
|
dclk1b
|
doublecortin-like kinase 1b |
| chr5_+_44316830 | 0.37 |
|
|
|
| chr5_-_30324182 | 0.37 |
ENSDART00000153909
|
spns2
|
spinster homolog 2 (Drosophila) |
| chr20_-_47123598 | 0.37 |
ENSDART00000100320
|
dnmt3aa
|
DNA (cytosine-5-)-methyltransferase 3 alpha a |
| chr6_+_170792 | 0.37 |
|
|
|
| chr8_+_10267246 | 0.37 |
ENSDART00000159312
ENSDART00000160766 |
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr2_-_42279180 | 0.37 |
ENSDART00000047055
|
trim55a
|
tripartite motif containing 55a |
| chr16_+_29277783 | 0.37 |
ENSDART00000149569
|
CR848047.1
|
ENSDARG00000095906 |
| chr1_+_4704661 | 0.36 |
ENSDART00000040204
|
tuba8l2
|
tubulin, alpha 8 like 2 |
| chr2_-_21323717 | 0.36 |
ENSDART00000113384
|
lyrm4
|
LYR motif containing 4 |
| chr11_-_2524674 | 0.36 |
ENSDART00000114079
|
nab2
|
NGFI-A binding protein 2 (EGR1 binding protein 2) |
| chr20_-_26142897 | 0.36 |
ENSDART00000140255
|
si:dkey-12h9.6
|
si:dkey-12h9.6 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.3 | GO:0042985 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.4 | 1.3 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.4 | 2.0 | GO:1901739 | regulation of myoblast fusion(GO:1901739) |
| 0.4 | 2.7 | GO:0006833 | water transport(GO:0006833) |
| 0.3 | 3.3 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.2 | 1.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.2 | 0.7 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.2 | 0.8 | GO:0086065 | atrial cardiac muscle cell action potential(GO:0086014) cell-cell signaling involved in cardiac conduction(GO:0086019) atrial cardiac muscle cell to AV node cell signaling(GO:0086026) cell communication involved in cardiac conduction(GO:0086065) atrial cardiac muscle cell to AV node cell communication(GO:0086066) |
| 0.2 | 0.6 | GO:0042762 | regulation of sulfur metabolic process(GO:0042762) |
| 0.2 | 0.6 | GO:0071831 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 0.2 | 0.8 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.2 | 1.8 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.2 | 1.2 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.2 | 0.7 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.2 | 1.3 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.1 | 0.7 | GO:0033151 | V(D)J recombination(GO:0033151) |
| 0.1 | 0.3 | GO:0035474 | selective angioblast sprouting(GO:0035474) |
| 0.1 | 0.3 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.1 | 0.4 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.1 | 1.4 | GO:0098508 | endothelial to hematopoietic transition(GO:0098508) |
| 0.1 | 0.4 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.1 | 0.5 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 0.1 | 0.5 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.1 | 0.9 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.8 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 0.6 | GO:0010990 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.6 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.1 | 0.3 | GO:0007508 | larval development(GO:0002164) larval heart development(GO:0007508) |
| 0.1 | 0.4 | GO:0010656 | negative regulation of muscle cell apoptotic process(GO:0010656) muscle cell apoptotic process(GO:0010657) striated muscle cell apoptotic process(GO:0010658) regulation of muscle cell apoptotic process(GO:0010660) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) |
| 0.1 | 0.4 | GO:0070296 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) sarcoplasmic reticulum calcium ion transport(GO:0070296) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.1 | 0.3 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.4 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 0.4 | GO:0018158 | peptidyl-lysine oxidation(GO:0018057) protein oxidation(GO:0018158) |
| 0.1 | 0.4 | GO:0090299 | regulation of neural crest formation(GO:0090299) |
| 0.1 | 3.6 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 0.4 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 0.8 | GO:2000177 | regulation of neural precursor cell proliferation(GO:2000177) |
| 0.1 | 0.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.3 | GO:0000050 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.1 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.9 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.1 | 0.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.1 | 0.4 | GO:1903069 | neuronal action potential(GO:0019228) regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903069) |
| 0.1 | 0.3 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 0.3 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.3 | GO:0035989 | tendon development(GO:0035989) |
| 0.1 | 0.9 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 0.3 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 0.1 | 0.3 | GO:1904969 | bleb assembly(GO:0032060) slow muscle cell migration(GO:1904969) |
| 0.1 | 0.2 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.1 | 0.5 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.2 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.5 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 1.0 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.1 | 0.4 | GO:0045453 | bone resorption(GO:0045453) |
| 0.1 | 0.3 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.1 | 1.1 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.1 | 0.2 | GO:1902895 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.1 | 0.9 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.1 | 1.0 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.1 | 1.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.3 | GO:0034650 | cortisol metabolic process(GO:0034650) |
| 0.1 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.3 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.1 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.1 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.4 | GO:2000480 | regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 0.3 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.1 | 1.0 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.1 | 0.2 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.1 | 0.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.6 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.6 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.2 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.0 | 0.2 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.3 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 0.4 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.0 | 0.2 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.0 | 0.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 1.1 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.2 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.4 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.1 | GO:0070166 | enamel mineralization(GO:0070166) amelogenesis(GO:0097186) |
| 0.0 | 0.5 | GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway(GO:0008277) |
| 0.0 | 0.5 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.0 | 0.6 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.2 | GO:0019878 | lysine biosynthetic process(GO:0009085) lysine biosynthetic process via aminoadipic acid(GO:0019878) |
| 0.0 | 0.4 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.3 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.7 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 1.1 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.2 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.7 | GO:0006482 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.0 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.0 | 0.3 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.3 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) |
| 0.0 | 0.2 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 1.6 | GO:1902668 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.2 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.4 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 1.6 | GO:0022904 | respiratory electron transport chain(GO:0022904) |
| 0.0 | 0.1 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.0 | 0.1 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 1.0 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 1.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.8 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.2 | GO:0002698 | negative regulation of immune effector process(GO:0002698) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.5 | GO:0061009 | intrahepatic bile duct development(GO:0035622) common bile duct development(GO:0061009) |
| 0.0 | 0.3 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.5 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.2 | GO:0001556 | oocyte maturation(GO:0001556) |
| 0.0 | 0.2 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.7 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.0 | 0.2 | GO:0001952 | regulation of cell-matrix adhesion(GO:0001952) |
| 0.0 | 0.1 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.0 | 1.0 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine diphosphate metabolic process(GO:0042357) thiamine-containing compound metabolic process(GO:0042723) |
| 0.0 | 0.2 | GO:0032261 | purine nucleotide salvage(GO:0032261) |
| 0.0 | 0.3 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.1 | GO:0032475 | otolith formation(GO:0032475) |
| 0.0 | 0.2 | GO:0061055 | myotome development(GO:0061055) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0071907 | determination of digestive tract left/right asymmetry(GO:0071907) |
| 0.0 | 0.0 | GO:2000136 | cell proliferation involved in heart morphogenesis(GO:0061323) regulation of cell proliferation involved in heart morphogenesis(GO:2000136) |
| 0.0 | 0.2 | GO:0006144 | purine nucleobase metabolic process(GO:0006144) |
| 0.0 | 0.1 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.1 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.2 | GO:0006188 | IMP biosynthetic process(GO:0006188) 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.3 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.0 | 0.2 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.5 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.1 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.0 | 0.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.0 | 0.6 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.0 | 0.2 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.2 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.4 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.0 | 0.5 | GO:0045765 | regulation of angiogenesis(GO:0045765) |
| 0.0 | 0.2 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) |
| 0.0 | 0.1 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 1.1 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 0.1 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.1 | GO:0003313 | heart rudiment development(GO:0003313) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.8 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.4 | GO:0001570 | vasculogenesis(GO:0001570) |
| 0.0 | 0.2 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.4 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 1.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.0 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.4 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.7 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.0 | 0.3 | GO:0030641 | regulation of cellular pH(GO:0030641) |
| 0.0 | 0.1 | GO:2000142 | regulation of transcription initiation from RNA polymerase II promoter(GO:0060260) regulation of DNA-templated transcription, initiation(GO:2000142) |
| 0.0 | 0.2 | GO:0042743 | hydrogen peroxide metabolic process(GO:0042743) |
| 0.0 | 0.2 | GO:0061386 | closure of optic fissure(GO:0061386) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 1.3 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 1.7 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.2 | 0.6 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.2 | 0.6 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.2 | 0.5 | GO:0008352 | katanin complex(GO:0008352) |
| 0.1 | 0.6 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 0.4 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.1 | 1.0 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.3 | GO:0031362 | intrinsic component of external side of plasma membrane(GO:0031233) anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.6 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.5 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.1 | 0.2 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.1 | 0.5 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.1 | 0.9 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.1 | 0.2 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.1 | 0.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.1 | 0.9 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.6 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.4 | GO:0098533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.1 | 3.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 0.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.7 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.7 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.9 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.8 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 1.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.2 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.4 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 2.1 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.3 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.0 | 1.5 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) cytochrome complex(GO:0070069) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 1.0 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.3 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.2 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 1.2 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.1 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 0.5 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 1.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.7 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 3.0 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 5.8 | GO:0030054 | cell junction(GO:0030054) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 7.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.5 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.0 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.0 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.0 | GO:1990072 | TRAPPIII protein complex(GO:1990072) |
| 0.0 | 0.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.4 | 2.7 | GO:0015250 | water channel activity(GO:0015250) |
| 0.4 | 1.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.3 | 3.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 0.6 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.2 | 1.8 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.2 | 1.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 0.5 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 2.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.8 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.4 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.1 | 0.4 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.1 | 0.5 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.3 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.6 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.1 | 0.9 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.1 | 0.4 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.1 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 1.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.2 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 1.2 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.8 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.1 | 0.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.2 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.5 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.9 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 0.2 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.1 | 0.4 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.2 | GO:0003920 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 1.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.4 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.9 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 1.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.4 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.5 | GO:0030291 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 2.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.4 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 1.5 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.5 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.9 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.6 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.4 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 2.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.6 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 1.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 1.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.3 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.2 | GO:0038064 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.2 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.5 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.2 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.1 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) |
| 0.0 | 0.1 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.2 | GO:0031704 | apelin receptor binding(GO:0031704) |
| 0.0 | 0.1 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.0 | 0.9 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.0 | 0.4 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.6 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.3 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 0.8 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.3 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.1 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.0 | 0.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.4 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 1.0 | GO:0016859 | cis-trans isomerase activity(GO:0016859) |
| 0.0 | 0.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.7 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 1.8 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.7 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 1.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 9.2 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.1 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.0 | GO:0004953 | icosanoid receptor activity(GO:0004953) |
| 0.0 | 0.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.3 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.0 | 0.5 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.3 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.2 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.2 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.0 | 0.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.8 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.8 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.1 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.1 | 4.1 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 1.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.4 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.7 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.7 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.6 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.4 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 0.9 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 0.3 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.1 | 0.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 2.7 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.1 | 0.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.1 | 0.9 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 1.1 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 2.4 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.3 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.0 | 0.3 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.5 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.3 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |