Project
DANIO-CODE
Navigation
Downloads
Results for smad3a+smad3b
Z-value: 0.78
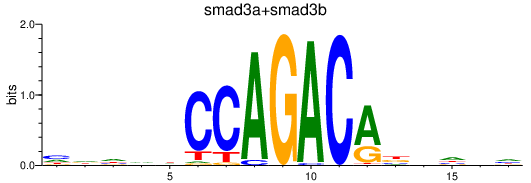
Transcription factors associated with smad3a+smad3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
smad3b
|
ENSDARG00000010207 | SMAD family member 3b |
|
smad3a
|
ENSDARG00000036096 | SMAD family member 3a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| smad3a | dr10_dc_chr7_-_33878076_33878147 | 0.40 | 1.3e-01 | Click! |
| smad3b | dr10_dc_chr18_+_19783732_19783788 | 0.38 | 1.5e-01 | Click! |
Activity profile of smad3a+smad3b motif
Sorted Z-values of smad3a+smad3b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of smad3a+smad3b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_23802054 | 2.14 |
ENSDART00000144226
|
ctsll
|
cathepsin L, like |
| chr2_-_20941256 | 1.88 |
ENSDART00000114199
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr2_-_59047356 | 1.67 |
|
|
|
| chr19_-_3226831 | 1.67 |
ENSDART00000145710
ENSDART00000110763 ENSDART00000074620 |
stm
|
starmaker |
| chr21_+_26035166 | 1.38 |
ENSDART00000134939
|
rpl23a
|
ribosomal protein L23a |
| chr8_-_13503775 | 1.36 |
ENSDART00000063834
|
ENSDARG00000043482
|
ENSDARG00000043482 |
| chr12_-_17357133 | 1.30 |
ENSDART00000079115
|
papss2b
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2b |
| chr3_-_44113070 | 1.25 |
ENSDART00000160717
|
znf750
|
zinc finger protein 750 |
| chr1_+_46487300 | 1.18 |
ENSDART00000158432
ENSDART00000074450 |
morc3b
|
MORC family CW-type zinc finger 3b |
| chr24_-_33377293 | 1.13 |
ENSDART00000124938
|
si:ch1073-406l10.2
|
si:ch1073-406l10.2 |
| chr14_+_38445969 | 1.11 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr6_-_40774050 | 1.10 |
ENSDART00000076200
|
h1fx
|
H1 histone family, member X |
| chr14_+_27936303 | 1.06 |
ENSDART00000003293
|
mid2
|
midline 2 |
| chr6_-_39311711 | 1.06 |
|
|
|
| chr9_+_3147601 | 1.06 |
ENSDART00000135619
|
ftr52p
|
finTRIM family, member 52, pseudogene |
| chr10_+_26020615 | 1.05 |
ENSDART00000128292
ENSDART00000170275 ENSDART00000166164 ENSDART00000108808 |
frem2a
|
Fras1 related extracellular matrix protein 2a |
| chr25_+_35796086 | 1.04 |
ENSDART00000112196
|
HIST2H4B
|
zgc:163040 |
| chr3_+_48362748 | 1.04 |
ENSDART00000157199
|
mkl2b
|
MKL/myocardin-like 2b |
| chr8_-_38168395 | 1.03 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr23_-_9556986 | 1.01 |
ENSDART00000139688
|
soga1
|
suppressor of glucose, autophagy associated 1 |
| chr4_+_29842350 | 0.99 |
|
|
|
| chr22_-_28922834 | 0.97 |
ENSDART00000104828
|
gtpbp2b
|
GTP binding protein 2b |
| chr25_+_35765309 | 0.94 |
ENSDART00000128547
|
hist1h4l
|
histone 1, H4, like |
| chr6_+_6333766 | 0.89 |
ENSDART00000140827
|
bcl11ab
|
B-cell CLL/lymphoma 11Ab |
| chr8_+_1122000 | 0.87 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr4_-_72067910 | 0.87 |
ENSDART00000170308
|
BX855614.3
|
ENSDARG00000101193 |
| chr6_-_50705420 | 0.83 |
ENSDART00000074100
|
osgn1
|
oxidative stress induced growth inhibitor 1 |
| chr23_-_46250905 | 0.83 |
ENSDART00000160110
|
tgm1l4
|
transglutaminase 1 like 4 |
| chr19_-_9043897 | 0.82 |
ENSDART00000104657
|
mrps21
|
mitochondrial ribosomal protein S21 |
| chr23_-_24755654 | 0.80 |
ENSDART00000104035
|
ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr7_-_69639922 | 0.79 |
|
|
|
| chr11_-_23079615 | 0.75 |
ENSDART00000125024
|
golt1a
|
golgi transport 1A |
| chr23_+_2597825 | 0.73 |
ENSDART00000159039
|
ENSDARG00000089986
|
ENSDARG00000089986 |
| KN149816v1_-_3337 | 0.72 |
|
|
|
| chr20_-_53271601 | 0.72 |
|
|
|
| KN150670v1_-_72156 | 0.68 |
|
|
|
| chr25_+_34560773 | 0.68 |
ENSDART00000157248
|
ENSDARG00000097103
|
ENSDARG00000097103 |
| chr21_-_26035057 | 0.66 |
ENSDART00000141382
|
rab34b
|
RAB34, member RAS oncogene family b |
| chr14_-_27754549 | 0.63 |
ENSDART00000135337
|
zgc:64189
|
zgc:64189 |
| chr12_-_3741925 | 0.62 |
ENSDART00000092983
|
ENSDARG00000063555
|
ENSDARG00000063555 |
| chr8_-_37011436 | 0.62 |
ENSDART00000139567
|
renbp
|
renin binding protein |
| chr7_-_57941497 | 0.61 |
ENSDART00000114008
|
unm_hu7910
|
un-named hu7910 |
| chr4_-_14330113 | 0.58 |
ENSDART00000091151
|
nell2b
|
neural EGFL like 2b |
| chr14_+_6239755 | 0.57 |
ENSDART00000097214
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr4_-_74907755 | 0.55 |
ENSDART00000132323
|
ftr51
|
finTRIM family, member 51 |
| chr13_+_1314139 | 0.55 |
|
|
|
| chr15_-_41777233 | 0.54 |
ENSDART00000154230
|
ftr90
|
finTRIM family, member 90 |
| chr18_-_36956268 | 0.54 |
|
|
|
| chr8_+_8673307 | 0.54 |
ENSDART00000110854
|
elk1
|
ELK1, member of ETS oncogene family |
| chr2_+_54911012 | 0.53 |
ENSDART00000027313
|
ndufv2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2 |
| chr10_+_29351676 | 0.52 |
ENSDART00000088973
|
sytl2a
|
synaptotagmin-like 2a |
| chr3_-_28741820 | 0.52 |
ENSDART00000138840
|
eef2kmt
|
eukaryotic elongation factor 2 lysine methyltransferase |
| chr2_-_37914464 | 0.51 |
ENSDART00000129852
|
hbl1
|
hexose-binding lectin 1 |
| chr7_-_6273677 | 0.50 |
ENSDART00000173419
|
si:ch73-368j24.1
|
si:ch73-368j24.1 |
| chr2_-_59293269 | 0.49 |
ENSDART00000148286
|
ftr39p
|
finTRIM family, member 39, pseudogene |
| chr25_+_34551602 | 0.49 |
ENSDART00000121825
|
hist1h4l
|
histone 1, H4, like |
| chr11_-_14943957 | 0.49 |
ENSDART00000162938
|
hug
|
HuG |
| chr20_+_46309812 | 0.47 |
ENSDART00000100532
|
stx7l
|
syntaxin 7-like |
| chr6_+_58704352 | 0.46 |
ENSDART00000172466
|
igsf8
|
immunoglobulin superfamily, member 8 |
| chr2_-_20941390 | 0.46 |
ENSDART00000114199
|
si:ch211-267e7.3
|
si:ch211-267e7.3 |
| chr5_-_57016269 | 0.45 |
ENSDART00000074264
|
crfb12
|
cytokine receptor family member B12 |
| chr3_-_40385307 | 0.43 |
|
|
|
| chr25_-_34642724 | 0.43 |
ENSDART00000121963
|
hist1h4l
|
histone 1, H4, like |
| chr9_-_16845777 | 0.43 |
ENSDART00000160273
|
ENSDARG00000102261
|
ENSDARG00000102261 |
| chr23_-_46251021 | 0.43 |
ENSDART00000160110
|
tgm1l4
|
transglutaminase 1 like 4 |
| chr14_+_8198019 | 0.42 |
ENSDART00000123364
|
vegfba
|
vascular endothelial growth factor Ba |
| KN150423v1_-_9765 | 0.42 |
|
|
|
| chr1_+_53384116 | 0.42 |
ENSDART00000149760
|
triobpa
|
TRIO and F-actin binding protein a |
| chr19_+_628049 | 0.41 |
ENSDART00000098884
ENSDART00000170433 |
tert
|
telomerase reverse transcriptase |
| chr20_+_19662825 | 0.41 |
ENSDART00000103798
|
nme6
|
NME/NM23 nucleoside diphosphate kinase 6 |
| chr5_+_35198499 | 0.40 |
|
|
|
| chr17_-_45403518 | 0.38 |
ENSDART00000156002
|
tmem206
|
transmembrane protein 206 |
| chr2_+_23038657 | 0.38 |
ENSDART00000089012
|
kif1ab
|
kinesin family member 1Ab |
| chr17_+_14878846 | 0.37 |
ENSDART00000010507
ENSDART00000131052 |
ptger2a
|
prostaglandin E receptor 2a (subtype EP2) |
| chr23_+_25381458 | 0.37 |
ENSDART00000140463
|
esyt1a
|
extended synaptotagmin-like protein 1a |
| chr7_-_24119645 | 0.36 |
ENSDART00000141769
|
ptgr1
|
prostaglandin reductase 1 |
| chr1_+_44414936 | 0.36 |
|
|
|
| chr7_+_21455482 | 0.35 |
|
|
|
| chr7_+_5837533 | 0.35 |
ENSDART00000110813
|
hist1h4l
|
histone 1, H4, like |
| chr25_-_22242337 | 0.35 |
|
|
|
| chr5_+_1164747 | 0.35 |
ENSDART00000129490
|
gb:bc139872
|
expressed sequence BC139872 |
| chr15_+_46129807 | 0.33 |
ENSDART00000153936
|
si:ch1073-340i21.2
|
si:ch1073-340i21.2 |
| chr21_-_17566461 | 0.32 |
ENSDART00000044078
ENSDART00000168495 ENSDART00000020048 |
gsna
|
gelsolin a |
| chr17_-_27254921 | 0.31 |
ENSDART00000077087
|
id3
|
inhibitor of DNA binding 3 |
| chr1_-_22170553 | 0.31 |
ENSDART00000139412
|
SMIM18
|
small integral membrane protein 18 |
| chr10_-_22881453 | 0.31 |
|
|
|
| chr1_+_46487378 | 0.31 |
ENSDART00000137448
|
morc3b
|
MORC family CW-type zinc finger 3b |
| chr13_+_19115 | 0.30 |
ENSDART00000176574
|
CABZ01089797.1
|
ENSDARG00000107538 |
| chr20_+_46969915 | 0.30 |
ENSDART00000158124
|
ENSDARG00000103380
|
ENSDARG00000103380 |
| chr1_+_23093475 | 0.30 |
|
|
|
| chr4_-_75690772 | 0.30 |
ENSDART00000174183
|
CABZ01063884.1
|
ENSDARG00000105697 |
| chr5_+_30725475 | 0.30 |
|
|
|
| chr4_-_14330048 | 0.29 |
ENSDART00000091151
|
nell2b
|
neural EGFL like 2b |
| chr5_+_20340093 | 0.29 |
ENSDART00000139260
|
limk2
|
LIM domain kinase 2 |
| chr7_+_73603232 | 0.29 |
ENSDART00000112685
|
hist1h4l
|
histone 1, H4, like |
| chr20_-_53271658 | 0.29 |
|
|
|
| chr9_+_24089828 | 0.28 |
ENSDART00000127859
|
trim63b
|
tripartite motif containing 63b |
| chr17_+_44666606 | 0.28 |
ENSDART00000155096
|
tmem63c
|
transmembrane protein 63C |
| chr14_+_20614087 | 0.28 |
ENSDART00000160318
|
lygl2
|
lysozyme g-like 2 |
| chr18_+_17622557 | 0.27 |
ENSDART00000046891
|
cetp
|
cholesteryl ester transfer protein, plasma |
| chr24_+_4402474 | 0.27 |
ENSDART00000066836
|
GJD4
|
gap junction protein delta 4 |
| chr12_-_16514995 | 0.26 |
ENSDART00000128811
|
si:dkey-239j18.3
|
si:dkey-239j18.3 |
| chr20_+_4840692 | 0.26 |
|
|
|
| chr13_+_36638875 | 0.26 |
ENSDART00000111832
|
atl1
|
atlastin GTPase 1 |
| chr25_-_20170375 | 0.25 |
ENSDART00000138763
|
dnajb9a
|
DnaJ (Hsp40) homolog, subfamily B, member 9a |
| chr23_+_35954132 | 0.25 |
ENSDART00000103149
|
hoxc13a
|
homeobox C13a |
| chr15_+_9096765 | 0.24 |
ENSDART00000080196
|
si:dkey-202g17.3
|
si:dkey-202g17.3 |
| chr16_-_2570425 | 0.24 |
|
|
|
| chr3_+_31793328 | 0.24 |
ENSDART00000127330
ENSDART00000126773 ENSDART00000055279 |
snrnp70
|
small nuclear ribonucleoprotein 70 (U1) |
| chr1_-_29562875 | 0.24 |
ENSDART00000125759
|
si:dkey-22o12.2
|
si:dkey-22o12.2 |
| chr17_-_53266905 | 0.24 |
|
|
|
| chr21_+_25496520 | 0.24 |
ENSDART00000134052
|
si:dkey-17e16.8
|
si:dkey-17e16.8 |
| chr14_-_6822151 | 0.23 |
ENSDART00000171311
|
si:ch73-43g23.1
|
si:ch73-43g23.1 |
| chr3_+_24059652 | 0.23 |
ENSDART00000034762
|
prr15la
|
proline rich 15-like a |
| chr8_-_18167468 | 0.23 |
ENSDART00000137005
|
elovl8b
|
ELOVL fatty acid elongase 8b |
| chr24_-_25189173 | 0.22 |
|
|
|
| chr19_-_6963103 | 0.21 |
|
|
|
| chr10_+_17634410 | 0.21 |
ENSDART00000168635
|
KCNN2
|
potassium calcium-activated channel subfamily N member 2 |
| chr25_+_17906515 | 0.20 |
|
|
|
| chr8_+_23172960 | 0.20 |
ENSDART00000028946
|
tpd52l2a
|
tumor protein D52-like 2a |
| chr8_+_39726489 | 0.19 |
ENSDART00000037914
|
cox6a1
|
cytochrome c oxidase subunit VIa polypeptide 1 |
| chr21_+_25496914 | 0.19 |
ENSDART00000134052
|
si:dkey-17e16.8
|
si:dkey-17e16.8 |
| chr25_-_35451402 | 0.19 |
ENSDART00000066987
|
mphosph6
|
M-phase phosphoprotein 6 |
| chr8_-_18167570 | 0.19 |
ENSDART00000142114
|
elovl8b
|
ELOVL fatty acid elongase 8b |
| chr25_+_30838768 | 0.17 |
|
|
|
| chr20_-_54364559 | 0.17 |
|
|
|
| chr16_-_48189682 | 0.17 |
|
|
|
| chr24_+_42193701 | 0.16 |
ENSDART00000067734
|
snx16
|
sorting nexin 16 |
| KN150423v1_-_9882 | 0.16 |
|
|
|
| chr25_-_17906739 | 0.16 |
ENSDART00000149696
|
cep290
|
centrosomal protein 290 |
| chr17_-_44895892 | 0.16 |
ENSDART00000123971
|
fam161b
|
family with sequence similarity 161, member B |
| chr23_+_14172256 | 0.15 |
ENSDART00000155326
|
si:dkey-90a13.10
|
si:dkey-90a13.10 |
| chr5_-_31116505 | 0.15 |
ENSDART00000131443
|
dpm2
|
dolichyl-phosphate mannosyltransferase polypeptide 2, regulatory subunit |
| chr3_-_4590689 | 0.14 |
ENSDART00000163052
ENSDART00000164569 |
ENSDARG00000098540
|
ENSDARG00000098540 |
| chr19_+_48166365 | 0.14 |
|
|
|
| chr25_+_17906724 | 0.14 |
|
|
|
| chr24_-_41917835 | 0.14 |
|
|
|
| chr3_+_36497377 | 0.13 |
|
|
|
| chr10_+_21760897 | 0.12 |
ENSDART00000176794
|
pcdh1gb9
|
protocadherin 1 gamma b 9 |
| chr7_+_59494488 | 0.12 |
|
|
|
| chr22_+_1509147 | 0.12 |
ENSDART00000164089
|
si:ch211-255f4.6
|
si:ch211-255f4.6 |
| chr2_+_35505943 | 0.10 |
ENSDART00000131248
|
CR855337.1
|
ENSDARG00000094295 |
| chr18_+_17564926 | 0.10 |
|
|
|
| chr19_+_9868101 | 0.09 |
ENSDART00000138310
|
cacng6a
|
calcium channel, voltage-dependent, gamma subunit 6a |
| chr24_-_42193714 | 0.09 |
|
|
|
| chr3_-_32457740 | 0.09 |
|
|
|
| chr23_+_25005457 | 0.09 |
ENSDART00000136162
|
klhl21
|
kelch-like family member 21 |
| chr6_+_19476202 | 0.09 |
ENSDART00000113911
|
micall1a
|
MICAL-like 1a |
| chr5_-_1164454 | 0.08 |
ENSDART00000054070
|
surf2
|
surfeit 2 |
| chr3_-_40385511 | 0.08 |
|
|
|
| chr8_-_3287944 | 0.06 |
ENSDART00000057874
|
FUT9 (1 of many)
|
fucosyltransferase 9 |
| chr5_-_336852 | 0.06 |
|
|
|
| chr5_+_62696901 | 0.06 |
ENSDART00000124616
|
rab14
|
RAB14, member RAS oncogene family |
| chr17_+_44896297 | 0.06 |
ENSDART00000133103
|
coq6
|
coenzyme Q6 monooxygenase |
| chr12_-_46943791 | 0.05 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr7_-_51210658 | 0.05 |
ENSDART00000174026
|
nhsl2
|
NHS-like 2 |
| chr25_+_35451459 | 0.05 |
|
|
|
| chr25_+_19889409 | 0.05 |
ENSDART00000178266
ENSDART00000121226 |
FO704882.1
|
ENSDARG00000085826 |
| chr9_-_23107452 | 0.05 |
ENSDART00000007392
|
arl5a
|
ADP-ribosylation factor-like 5A |
| chr24_-_25189252 | 0.05 |
|
|
|
| chr12_-_18438618 | 0.03 |
ENSDART00000078172
|
bricd5
|
BRICHOS domain containing 5 |
| chr16_-_24646282 | 0.03 |
ENSDART00000126274
|
si:ch211-79k12.2
|
si:ch211-79k12.2 |
| chr7_-_2230502 | 0.03 |
ENSDART00000173879
|
si:cabz01007794.1
|
si:cabz01007794.1 |
| chr17_-_53266811 | 0.02 |
|
|
|
| chr25_+_35451721 | 0.01 |
|
|
|
| chr18_-_20570874 | 0.01 |
ENSDART00000141367
|
si:ch211-238n5.4
|
si:ch211-238n5.4 |
| chr16_-_32250329 | 0.00 |
ENSDART00000143928
|
rwdd1
|
RWD domain containing 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.2 | 1.3 | GO:0050427 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 | 0.6 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.2 | 0.7 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.1 | 1.3 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.1 | 1.1 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.1 | 0.3 | GO:0071554 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.1 | 0.3 | GO:0055091 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) reverse cholesterol transport(GO:0043691) phospholipid homeostasis(GO:0055091) |
| 0.1 | 0.4 | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading(GO:1900026) |
| 0.1 | 1.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.4 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.1 | 0.4 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.6 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.1 | 0.4 | GO:0046051 | UTP biosynthetic process(GO:0006228) UTP metabolic process(GO:0046051) |
| 0.0 | 1.1 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.9 | GO:0050773 | regulation of dendrite development(GO:0050773) |
| 0.0 | 0.1 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 2.5 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.6 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.2 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 0.2 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.2 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.2 | GO:0098927 | protein targeting to lysosome(GO:0006622) early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 1.1 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.3 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 1.1 | GO:0033334 | fin morphogenesis(GO:0033334) |
| 0.0 | 2.3 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.4 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.4 | GO:0007596 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.0 | 1.3 | GO:0008544 | epidermis development(GO:0008544) |
| 0.0 | 1.0 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.3 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 2.4 | GO:0006955 | immune response(GO:0006955) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) |
| 0.0 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.3 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 1.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 3.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.5 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 2.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.2 | GO:0005685 | U1 snRNP(GO:0005685) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0005461 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 0.2 | 1.3 | GO:0004779 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 1.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 0.4 | GO:0004953 | icosanoid receptor activity(GO:0004953) |
| 0.1 | 0.4 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.3 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.4 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 0.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.1 | 0.8 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.6 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 1.0 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 1.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 2.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.4 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.3 | GO:0031210 | phosphatidylcholine binding(GO:0031210) |
| 0.0 | 0.4 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 3.0 | GO:0046982 | protein heterodimerization activity(GO:0046982) |
| 0.0 | 0.4 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.6 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.5 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.9 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.5 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.3 | REACTOME GAP JUNCTION TRAFFICKING | Genes involved in Gap junction trafficking |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 1.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.4 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |