Project
DANIO-CODE
Navigation
Downloads
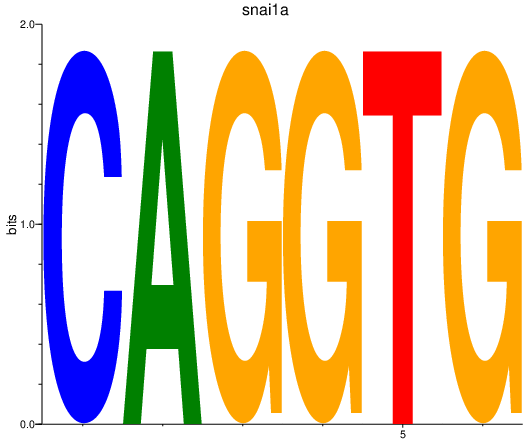
Results for snai1a
Z-value: 3.51
Transcription factors associated with snai1a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
snai1a
|
ENSDARG00000056995 | snail family zinc finger 1a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| snai1a | dr10_dc_chr11_-_25019899_25019981 | 0.64 | 7.8e-03 | Click! |
Activity profile of snai1a motif
Sorted Z-values of snai1a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of snai1a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_67475765 | 8.79 |
ENSDART00000160086
|
zgc:162592
|
zgc:162592 |
| chr23_+_540624 | 7.72 |
ENSDART00000034707
|
lsm14b
|
LSM family member 14B |
| chr11_+_30005768 | 7.24 |
ENSDART00000167618
|
CR790368.1
|
ENSDARG00000100936 |
| chr12_-_4264663 | 7.21 |
ENSDART00000152521
|
ca15b
|
carbonic anhydrase XVb |
| chr1_-_44892852 | 6.48 |
ENSDART00000160961
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr1_-_44892600 | 6.34 |
ENSDART00000149155
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr23_+_2786407 | 6.34 |
ENSDART00000066086
|
zgc:114123
|
zgc:114123 |
| chr12_+_30389706 | 6.04 |
|
|
|
| chr7_-_55346967 | 6.00 |
ENSDART00000135304
|
pabpn1l
|
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
| chr21_-_804453 | 5.86 |
|
|
|
| chr6_+_135002 | 5.85 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr14_+_14850200 | 5.47 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr17_+_699708 | 5.42 |
ENSDART00000165144
|
siva1
|
SIVA1, apoptosis-inducing factor |
| chr16_-_25318413 | 5.38 |
ENSDART00000058943
|
ENSDARG00000040280
|
ENSDARG00000040280 |
| chr12_+_13704712 | 5.35 |
ENSDART00000152257
|
ppp1r16a
|
protein phosphatase 1, regulatory subunit 16A |
| chr14_+_29945327 | 5.33 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr5_-_14993912 | 5.25 |
ENSDART00000085943
|
taok3a
|
TAO kinase 3a |
| chr7_-_19116999 | 5.15 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr3_-_32699672 | 4.96 |
|
|
|
| chr13_-_21541531 | 4.90 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr6_+_40525779 | 4.86 |
ENSDART00000033819
|
prkcda
|
protein kinase C, delta a |
| chr19_+_33552393 | 4.81 |
ENSDART00000043039
|
fam84b
|
family with sequence similarity 84, member B |
| chr3_+_32285237 | 4.80 |
ENSDART00000157324
|
prrg2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr7_+_24257251 | 4.80 |
ENSDART00000136473
|
ENSDARG00000079281
|
ENSDARG00000079281 |
| chr12_+_13053552 | 4.75 |
ENSDART00000124799
|
si:ch211-103b1.2
|
si:ch211-103b1.2 |
| chr9_-_28444272 | 4.71 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr21_-_30256477 | 4.62 |
ENSDART00000137193
|
slbp2
|
stem-loop binding protein 2 |
| chr15_+_44168257 | 4.53 |
ENSDART00000176254
|
CU655961.7
|
ENSDARG00000106724 |
| chr22_-_22315679 | 4.49 |
ENSDART00000033479
|
si:ch211-129c21.1
|
si:ch211-129c21.1 |
| chr5_+_57254393 | 4.43 |
ENSDART00000050949
|
btg4
|
B-cell translocation gene 4 |
| chr7_+_58448953 | 4.42 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr11_-_13094557 | 4.30 |
ENSDART00000160989
|
elovl1b
|
ELOVL fatty acid elongase 1b |
| chr14_+_22159893 | 4.16 |
ENSDART00000019296
|
gdf9
|
growth differentiation factor 9 |
| chr14_-_47210912 | 4.16 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr9_+_38832959 | 4.13 |
ENSDART00000110651
|
slc12a8
|
solute carrier family 12, member 8 |
| chr5_-_32636372 | 4.13 |
ENSDART00000085512
ENSDART00000144694 |
kank1b
|
KN motif and ankyrin repeat domains 1b |
| chr16_+_39209567 | 4.13 |
ENSDART00000121756
|
sybu
|
syntabulin (syntaxin-interacting) |
| chr9_-_28444024 | 4.12 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr21_-_34227158 | 4.11 |
ENSDART00000169218
ENSDART00000064320 ENSDART00000172381 |
alg13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr16_-_25317921 | 4.07 |
|
|
|
| chr14_+_24543399 | 4.06 |
ENSDART00000106039
|
arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr13_-_36400356 | 4.05 |
ENSDART00000175311
|
pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr14_-_880799 | 4.04 |
ENSDART00000031992
|
rgs14a
|
regulator of G protein signaling 14a |
| chr15_-_19192862 | 3.96 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr2_+_28199458 | 3.96 |
ENSDART00000150330
|
buc
|
bucky ball |
| chr12_+_22552867 | 3.95 |
ENSDART00000152930
|
cdca9
|
cell division cycle associated 9 |
| chr18_-_25784873 | 3.93 |
ENSDART00000103046
|
zgc:162879
|
zgc:162879 |
| chr19_-_18664720 | 3.92 |
ENSDART00000108627
|
snx10a
|
sorting nexin 10a |
| chr1_-_44892558 | 3.82 |
ENSDART00000149155
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr10_-_25246786 | 3.81 |
ENSDART00000036906
|
kpna7
|
karyopherin alpha 7 (importin alpha 8) |
| chr17_+_25313170 | 3.77 |
ENSDART00000157309
|
tmem54a
|
transmembrane protein 54a |
| chr12_-_10474942 | 3.75 |
ENSDART00000106163
ENSDART00000124562 |
zgc:152977
|
zgc:152977 |
| chr6_+_10098305 | 3.75 |
ENSDART00000151477
|
cobll1a
|
cordon-bleu WH2 repeat protein-like 1a |
| chr15_+_29092022 | 3.75 |
ENSDART00000141164
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr20_-_14218080 | 3.70 |
ENSDART00000104032
|
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr20_-_43846604 | 3.70 |
ENSDART00000150078
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr14_-_7102535 | 3.69 |
ENSDART00000036463
|
dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr6_+_135255 | 3.68 |
ENSDART00000097468
|
zglp1
|
zinc finger, GATA-like protein 1 |
| chr9_+_8418408 | 3.66 |
ENSDART00000043067
|
zgc:171776
|
zgc:171776 |
| chr22_+_22413803 | 3.64 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr14_-_32937496 | 3.63 |
ENSDART00000048130
|
stard14
|
START domain containing 14 |
| chr2_-_4998074 | 3.62 |
|
|
|
| chr12_+_13704914 | 3.60 |
ENSDART00000152257
|
ppp1r16a
|
protein phosphatase 1, regulatory subunit 16A |
| chr3_+_59972297 | 3.60 |
|
|
|
| chr1_-_48876924 | 3.60 |
ENSDART00000143474
|
zp3c
|
zona pellucida glycoprotein 3c |
| chr19_+_29750506 | 3.59 |
ENSDART00000164432
|
ldlrap1a
|
low density lipoprotein receptor adaptor protein 1a |
| chr19_-_27093095 | 3.59 |
|
|
|
| chr15_-_43283562 | 3.53 |
ENSDART00000110352
|
ap1s3a
|
adaptor-related protein complex 1, sigma 3 subunit, a |
| KN150456v1_-_19515 | 3.49 |
ENSDART00000168786
|
h1m
|
linker histone H1M |
| chr24_-_38195383 | 3.47 |
ENSDART00000056381
|
crp2
|
C-reactive protein 2 |
| chr20_-_25587274 | 3.40 |
ENSDART00000141340
|
si:dkey-183n20.15
|
si:dkey-183n20.15 |
| chr10_+_43953171 | 3.39 |
|
|
|
| chr12_-_42212994 | 3.39 |
ENSDART00000171075
|
zgc:111868
|
zgc:111868 |
| chr14_+_20045326 | 3.38 |
ENSDART00000167637
|
aff2
|
AF4/FMR2 family, member 2 |
| chr17_+_25314229 | 3.35 |
ENSDART00000082319
|
tmem54a
|
transmembrane protein 54a |
| chr18_-_40718244 | 3.32 |
ENSDART00000077577
|
si:ch211-132b12.8
|
si:ch211-132b12.8 |
| chr11_-_26819599 | 3.32 |
ENSDART00000043091
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr18_-_26799173 | 3.29 |
ENSDART00000136776
ENSDART00000076484 |
kti12
|
KTI12 chromatin associated homolog |
| chr19_+_15536640 | 3.27 |
ENSDART00000098970
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr15_-_35020312 | 3.22 |
ENSDART00000154094
|
mgat1a
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase a |
| chr17_-_38939792 | 3.20 |
ENSDART00000141177
|
slc24a4a
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4a |
| chr15_+_29092224 | 3.18 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr14_-_33641145 | 3.18 |
ENSDART00000167774
|
foxa
|
forkhead box A sequence |
| chr17_-_36913213 | 3.17 |
ENSDART00000154981
|
senp6b
|
SUMO1/sentrin specific peptidase 6b |
| chr4_-_2347573 | 3.14 |
ENSDART00000149330
|
kcnc2
|
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr13_-_37530793 | 3.14 |
ENSDART00000141295
|
si:dkey-188i13.11
|
si:dkey-188i13.11 |
| chr11_+_29299382 | 3.11 |
|
|
|
| chr16_-_44179964 | 3.10 |
ENSDART00000058685
|
zfpm2a
|
zinc finger protein, FOG family member 2a |
| chr2_-_47766563 | 3.10 |
ENSDART00000038228
|
ap1s3b
|
adaptor-related protein complex 1, sigma 3 subunit, b |
| chr16_+_19831573 | 3.06 |
ENSDART00000135359
|
macc1
|
metastasis associated in colon cancer 1 |
| chr7_-_51497945 | 3.06 |
ENSDART00000054591
|
bmp15
|
bone morphogenetic protein 15 |
| KN150589v1_-_5209 | 3.02 |
ENSDART00000157761
ENSDART00000157531 |
elovl7b
|
ELOVL fatty acid elongase 7b |
| chr16_-_42116471 | 3.01 |
ENSDART00000058620
|
zp3d.1
|
zona pellucida glycoprotein 3d tandem duplicate 1 |
| chr15_-_25333773 | 3.01 |
ENSDART00000078230
|
mettl16
|
methyltransferase like 16 |
| chr12_-_4264610 | 3.01 |
ENSDART00000152377
|
ca15b
|
carbonic anhydrase XVb |
| chr2_+_7014241 | 3.00 |
ENSDART00000016607
|
rgs5b
|
regulator of G protein signaling 5b |
| chr15_-_19192933 | 2.99 |
ENSDART00000152428
|
arhgap32a
|
Rho GTPase activating protein 32a |
| chr22_+_24112851 | 2.98 |
|
|
|
| chr7_-_47990610 | 2.98 |
ENSDART00000147968
|
rbpms2b
|
RNA binding protein with multiple splicing 2b |
| chr3_-_31747486 | 2.98 |
ENSDART00000076189
|
zgc:171779
|
zgc:171779 |
| chr19_+_2942485 | 2.96 |
ENSDART00000177848
|
CABZ01066434.1
|
ENSDARG00000107451 |
| chr13_+_31586338 | 2.95 |
ENSDART00000034745
|
prkcha
|
protein kinase C, eta, a |
| chr9_-_8318363 | 2.91 |
ENSDART00000139867
|
si:ch211-145c1.1
|
si:ch211-145c1.1 |
| chr15_+_38397772 | 2.90 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr25_+_16098033 | 2.89 |
ENSDART00000141994
|
mical2b
|
microtubule associated monooxygenase, calponin and LIM domain containing 2b |
| chr14_-_35932521 | 2.87 |
ENSDART00000158722
|
BX511223.1
|
ENSDARG00000101064 |
| chr5_-_31738565 | 2.86 |
ENSDART00000017956
|
dab2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr3_+_32617947 | 2.84 |
ENSDART00000125260
|
hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase |
| chr2_-_30216377 | 2.83 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr1_-_18118467 | 2.82 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr8_-_3974320 | 2.82 |
ENSDART00000159895
|
mtmr3
|
myotubularin related protein 3 |
| chr12_-_23244600 | 2.82 |
ENSDART00000170376
|
mpp7a
|
membrane protein, palmitoylated 7a (MAGUK p55 subfamily member 7) |
| chr3_+_42217236 | 2.80 |
ENSDART00000168228
|
tmem184a
|
transmembrane protein 184a |
| chr10_-_7513764 | 2.80 |
ENSDART00000167054
ENSDART00000167706 |
nrg1
|
neuregulin 1 |
| chr16_+_35952050 | 2.80 |
ENSDART00000162411
|
sh3d21
|
SH3 domain containing 21 |
| chr7_-_47978449 | 2.80 |
ENSDART00000127007
ENSDART00000024062 |
cpeb1b
|
cytoplasmic polyadenylation element binding protein 1b |
| chr16_-_25318260 | 2.80 |
ENSDART00000058943
|
ENSDARG00000040280
|
ENSDARG00000040280 |
| chr7_-_26226451 | 2.77 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr1_-_44893257 | 2.76 |
ENSDART00000160961
|
atf7ip
|
activating transcription factor 7 interacting protein |
| KN150623v1_+_258 | 2.75 |
|
|
|
| chr5_+_3683209 | 2.75 |
ENSDART00000132056
|
ggnbp2
|
gametogenetin binding protein 2 |
| chr21_-_38979792 | 2.74 |
ENSDART00000056878
|
traf4b
|
tnf receptor-associated factor 4b |
| chr1_-_52833379 | 2.73 |
ENSDART00000143349
|
zgc:66455
|
zgc:66455 |
| chr23_+_540664 | 2.73 |
ENSDART00000034707
|
lsm14b
|
LSM family member 14B |
| chr20_+_4034072 | 2.71 |
ENSDART00000092217
|
ttc13
|
tetratricopeptide repeat domain 13 |
| chr22_+_25734113 | 2.71 |
ENSDART00000136334
|
si:ch211-250e5.16
|
si:ch211-250e5.16 |
| chr2_-_30216333 | 2.66 |
ENSDART00000130142
|
ube2w
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr23_+_39961676 | 2.65 |
ENSDART00000161881
|
ENSDARG00000104435
|
ENSDARG00000104435 |
| chr16_-_25318334 | 2.65 |
ENSDART00000058943
|
ENSDARG00000040280
|
ENSDARG00000040280 |
| chr2_+_32863386 | 2.65 |
ENSDART00000056649
|
tmem53
|
transmembrane protein 53 |
| chr10_-_2915710 | 2.65 |
ENSDART00000002622
|
oclna
|
occludin a |
| chr11_-_6442588 | 2.62 |
ENSDART00000137879
|
zgc:162969
|
zgc:162969 |
| chr4_+_14983045 | 2.62 |
ENSDART00000067046
|
cax1
|
cation/H+ exchanger protein 1 |
| chr22_+_349237 | 2.61 |
|
|
|
| chr12_+_22551105 | 2.59 |
ENSDART00000123808
ENSDART00000159864 |
cdca9
|
cell division cycle associated 9 |
| chr7_+_69211965 | 2.59 |
ENSDART00000028064
|
ctdnep1b
|
CTD nuclear envelope phosphatase 1b |
| chr14_+_22160235 | 2.58 |
ENSDART00000019296
|
gdf9
|
growth differentiation factor 9 |
| chr2_+_9762781 | 2.58 |
ENSDART00000003465
|
gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr17_-_30635487 | 2.57 |
ENSDART00000154960
|
sh3yl1
|
SH3 and SYLF domain containing 1 |
| chr10_+_19625897 | 2.57 |
|
|
|
| chr14_+_44703094 | 2.57 |
|
|
|
| chr23_-_37553481 | 2.57 |
|
|
|
| chr22_+_25113293 | 2.55 |
ENSDART00000171851
|
si:ch211-226h8.4
|
si:ch211-226h8.4 |
| chr12_+_47448318 | 2.54 |
ENSDART00000152857
|
fmn2b
|
formin 2b |
| chr8_-_37989930 | 2.52 |
|
|
|
| chr14_+_27984076 | 2.52 |
ENSDART00000173237
|
mid2
|
midline 2 |
| chr1_+_45784384 | 2.52 |
|
|
|
| chr8_-_26210618 | 2.52 |
ENSDART00000133466
|
CR847503.1
|
ENSDARG00000092715 |
| chr1_+_44516337 | 2.52 |
ENSDART00000125037
|
EVI5L
|
ecotropic viral integration site 5 like |
| chr14_+_25973961 | 2.51 |
ENSDART00000178140
ENSDART00000179471 |
FO082779.2
|
ENSDARG00000106651 |
| chr8_+_7625846 | 2.51 |
ENSDART00000175045
|
CABZ01078218.1
|
ENSDARG00000107577 |
| chr20_+_14218237 | 2.50 |
ENSDART00000044937
|
kcns3b
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3b |
| chr15_+_29091983 | 2.50 |
ENSDART00000141164
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr2_+_9762627 | 2.50 |
ENSDART00000003465
|
gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr13_-_28474489 | 2.50 |
|
|
|
| chr19_-_7402246 | 2.48 |
ENSDART00000092375
|
oxr1b
|
oxidation resistance 1b |
| chr19_-_33264349 | 2.47 |
|
|
|
| chr8_-_17736169 | 2.46 |
ENSDART00000063592
ENSDART00000175064 |
prkcz
|
protein kinase C, zeta |
| chr2_-_21512154 | 2.46 |
ENSDART00000135417
|
bmi1b
|
bmi1 polycomb ring finger oncogene 1b |
| chr12_+_20569659 | 2.46 |
ENSDART00000141804
|
st6galnac1.2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1, tandem duplicate 2 |
| chr17_-_25313024 | 2.46 |
ENSDART00000082324
|
zpcx
|
zona pellucida protein C |
| chr7_+_39408883 | 2.45 |
ENSDART00000133420
|
tbc1d14
|
TBC1 domain family, member 14 |
| chr2_+_44659334 | 2.45 |
ENSDART00000155017
|
pask
|
PAS domain containing serine/threonine kinase |
| chr19_+_33392145 | 2.45 |
ENSDART00000171782
|
spire1a
|
spire-type actin nucleation factor 1a |
| chr9_-_34459799 | 2.43 |
ENSDART00000059955
|
ildr1b
|
immunoglobulin-like domain containing receptor 1b |
| chr25_-_12148502 | 2.43 |
|
|
|
| chr5_+_58009274 | 2.42 |
ENSDART00000019561
|
zgc:171734
|
zgc:171734 |
| chr8_-_37989317 | 2.42 |
ENSDART00000067809
|
rab11fip1a
|
RAB11 family interacting protein 1 (class I) a |
| chr4_+_9278836 | 2.42 |
ENSDART00000014897
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr12_-_292556 | 2.39 |
ENSDART00000152608
|
CU611036.1
|
ENSDARG00000079888 |
| chr15_+_38397715 | 2.38 |
ENSDART00000142403
|
si:dkey-24p1.6
|
si:dkey-24p1.6 |
| chr7_+_58449163 | 2.38 |
ENSDART00000167166
|
zgc:56231
|
zgc:56231 |
| chr10_-_25808033 | 2.38 |
ENSDART00000134176
|
postna
|
periostin, osteoblast specific factor a |
| chr22_-_17627900 | 2.36 |
ENSDART00000138483
|
si:ch73-243b8.4
|
si:ch73-243b8.4 |
| chr5_-_15971338 | 2.35 |
ENSDART00000110437
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr25_+_5845303 | 2.35 |
ENSDART00000163948
|
ENSDARG00000053246
|
ENSDARG00000053246 |
| chr5_-_67121850 | 2.34 |
ENSDART00000017881
|
eif4e1b
|
eukaryotic translation initiation factor 4E family member 1B |
| chr1_-_33717934 | 2.34 |
ENSDART00000083736
|
lmo7b
|
LIM domain 7b |
| chr5_-_48013093 | 2.33 |
ENSDART00000083229
|
mblac2
|
metallo-beta-lactamase domain containing 2 |
| chr22_+_17803309 | 2.33 |
ENSDART00000136016
|
hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr24_+_26223767 | 2.33 |
|
|
|
| chr9_+_28420988 | 2.33 |
|
|
|
| chr12_+_21176669 | 2.31 |
ENSDART00000178562
|
ca10a
|
carbonic anhydrase Xa |
| chr3_+_42395984 | 2.31 |
ENSDART00000162096
|
micall2a
|
mical-like 2a |
| chr4_-_4498775 | 2.31 |
ENSDART00000130588
|
tbc1d30
|
TBC1 domain family, member 30 |
| chr19_-_20819101 | 2.31 |
ENSDART00000137590
|
dazl
|
deleted in azoospermia-like |
| chr19_-_307085 | 2.30 |
ENSDART00000145916
|
lingo4a
|
leucine rich repeat and Ig domain containing 4a |
| chr24_+_34183462 | 2.30 |
ENSDART00000143995
|
zgc:92591
|
zgc:92591 |
| chr9_-_28443972 | 2.30 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr5_-_8206288 | 2.29 |
ENSDART00000062957
|
paip1
|
poly(A) binding protein interacting protein 1 |
| chr7_+_67475893 | 2.27 |
ENSDART00000160086
|
zgc:162592
|
zgc:162592 |
| chr22_-_24964889 | 2.27 |
ENSDART00000102751
|
si:dkey-179j5.5
|
si:dkey-179j5.5 |
| chr12_-_17370920 | 2.26 |
ENSDART00000130735
|
minpp1b
|
multiple inositol-polyphosphate phosphatase 1b |
| chr23_+_6652454 | 2.26 |
ENSDART00000081763
|
rbm38
|
RNA binding motif protein 38 |
| chr13_-_33527029 | 2.26 |
|
|
|
| chr13_+_34563757 | 2.26 |
ENSDART00000133661
|
tasp1
|
taspase, threonine aspartase, 1 |
| chr5_-_11530803 | 2.24 |
ENSDART00000159896
|
gatsl3
|
GATS protein-like 3 |
| chr23_+_29017954 | 2.24 |
ENSDART00000140291
|
CR677513.1
|
ENSDARG00000093890 |
| chr4_+_90885 | 2.24 |
ENSDART00000169805
|
eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr11_-_44734403 | 2.23 |
ENSDART00000160656
|
afmid
|
arylformamidase |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 9.4 | GO:0040038 | meiotic cytokinesis(GO:0033206) polar body extrusion after meiotic divisions(GO:0040038) |
| 1.8 | 12.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 1.7 | 20.7 | GO:0090309 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 1.5 | 7.6 | GO:0060965 | negative regulation of posttranscriptional gene silencing(GO:0060149) negative regulation of gene silencing by miRNA(GO:0060965) negative regulation of gene silencing by RNA(GO:0060967) |
| 1.5 | 9.1 | GO:0030237 | female sex determination(GO:0030237) |
| 1.4 | 4.2 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 1.3 | 6.6 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 1.3 | 4.0 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 1.3 | 9.1 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 1.2 | 4.8 | GO:0051151 | regulation of smooth muscle cell differentiation(GO:0051150) negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 1.2 | 10.5 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 1.1 | 4.2 | GO:0043622 | cortical microtubule organization(GO:0043622) |
| 1.1 | 17.9 | GO:0060046 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 1.0 | 6.2 | GO:0010693 | regulation of alkaline phosphatase activity(GO:0010692) negative regulation of alkaline phosphatase activity(GO:0010693) |
| 1.0 | 3.0 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.9 | 5.5 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.9 | 6.5 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.9 | 2.7 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.8 | 2.5 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.8 | 5.8 | GO:0008585 | female gonad development(GO:0008585) |
| 0.8 | 0.8 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.8 | 3.1 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.7 | 19.7 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.7 | 0.7 | GO:0045777 | positive regulation of blood pressure(GO:0045777) |
| 0.7 | 2.9 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.7 | 7.2 | GO:0031111 | negative regulation of microtubule depolymerization(GO:0007026) negative regulation of microtubule polymerization or depolymerization(GO:0031111) |
| 0.7 | 2.8 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.7 | 2.1 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.7 | 2.1 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.7 | 0.7 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.7 | 3.4 | GO:1901841 | regulation of high voltage-gated calcium channel activity(GO:1901841) negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.7 | 2.0 | GO:2000725 | regulation of cardiocyte differentiation(GO:1905207) regulation of cardiac muscle cell differentiation(GO:2000725) |
| 0.7 | 2.0 | GO:0007344 | karyogamy(GO:0000741) pronuclear fusion(GO:0007344) |
| 0.7 | 2.7 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.7 | 4.0 | GO:0034036 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.7 | 3.9 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.6 | 1.9 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.6 | 1.9 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.6 | 8.6 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.6 | 13.5 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.6 | 6.1 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.6 | 1.7 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.6 | 6.9 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.6 | 6.8 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.6 | 2.2 | GO:0010759 | positive regulation of lamellipodium assembly(GO:0010592) regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) skeletal muscle satellite cell proliferation(GO:0014841) regulation of skeletal muscle satellite cell proliferation(GO:0014842) negative regulation of skeletal muscle cell proliferation(GO:0014859) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) positive regulation of lamellipodium organization(GO:1902745) |
| 0.5 | 2.7 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.5 | 4.4 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.5 | 4.8 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.5 | 2.7 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.5 | 7.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.5 | 1.5 | GO:0070657 | mechanosensory epithelium regeneration(GO:0070655) mechanoreceptor differentiation involved in mechanosensory epithelium regeneration(GO:0070656) neuromast regeneration(GO:0070657) neuromast hair cell differentiation involved in neuromast regeneration(GO:0070658) |
| 0.5 | 1.5 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.5 | 2.4 | GO:1901255 | nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.5 | 4.8 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.5 | 1.9 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.5 | 18.4 | GO:0048599 | oocyte development(GO:0048599) |
| 0.5 | 1.4 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) cellular response to vitamin D(GO:0071305) |
| 0.5 | 2.3 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.5 | 1.4 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.5 | 2.3 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.5 | 3.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.4 | 3.6 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.4 | 8.8 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.4 | 1.3 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.4 | 1.7 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.4 | 0.8 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.4 | 2.5 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.4 | 3.8 | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) |
| 0.4 | 1.3 | GO:0000493 | box H/ACA snoRNP assembly(GO:0000493) |
| 0.4 | 1.3 | GO:0046080 | dUTP metabolic process(GO:0046080) dUTP catabolic process(GO:0046081) |
| 0.4 | 0.8 | GO:0035264 | multicellular organism growth(GO:0035264) |
| 0.4 | 5.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.4 | 1.6 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.4 | 4.0 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.4 | 1.2 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.4 | 1.2 | GO:0045046 | peroxisomal membrane transport(GO:0015919) protein import into peroxisome membrane(GO:0045046) |
| 0.4 | 1.5 | GO:0010916 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) regulation of lipoprotein particle clearance(GO:0010984) negative regulation of lipoprotein particle clearance(GO:0010985) |
| 0.4 | 1.2 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.4 | 1.1 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.4 | 2.3 | GO:0032366 | intracellular sterol transport(GO:0032366) |
| 0.4 | 2.3 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.4 | 2.6 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.4 | 0.4 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.4 | 2.2 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.4 | 2.2 | GO:1905066 | regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.4 | 1.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.4 | 1.8 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) |
| 0.4 | 2.5 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.4 | 1.4 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.4 | 1.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.3 | 1.0 | GO:0098743 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.3 | 0.7 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.3 | 1.0 | GO:0098751 | bone cell development(GO:0098751) |
| 0.3 | 1.9 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.3 | 1.0 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.3 | 1.9 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.3 | 0.9 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.3 | 3.5 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.3 | 0.9 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine diphosphate metabolic process(GO:0042357) thiamine-containing compound metabolic process(GO:0042723) |
| 0.3 | 1.2 | GO:0051204 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.3 | 1.2 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.3 | 1.5 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.3 | 3.9 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.3 | 1.8 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.3 | 4.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 4.4 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.3 | 1.5 | GO:0022615 | protein import into peroxisome matrix, docking(GO:0016560) protein to membrane docking(GO:0022615) |
| 0.3 | 1.2 | GO:0015785 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.3 | 3.7 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.3 | 1.1 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.3 | 4.6 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.3 | 0.8 | GO:0035552 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded DNA demethylation(GO:0035552) |
| 0.3 | 2.7 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.3 | 2.6 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.3 | 0.8 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.3 | 1.3 | GO:0009110 | vitamin biosynthetic process(GO:0009110) |
| 0.3 | 3.8 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.3 | 1.0 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.2 | 0.5 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.2 | 1.2 | GO:0043545 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.2 | 1.0 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.2 | 2.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.2 | 1.0 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.2 | 1.4 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.2 | 2.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.2 | 1.2 | GO:0072319 | clathrin coat disassembly(GO:0072318) vesicle uncoating(GO:0072319) |
| 0.2 | 0.9 | GO:0003400 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.2 | 12.1 | GO:0035303 | regulation of dephosphorylation(GO:0035303) |
| 0.2 | 3.5 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.2 | 1.2 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.2 | 2.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.2 | 6.7 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.2 | 1.1 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.2 | 0.9 | GO:0007032 | endosome organization(GO:0007032) |
| 0.2 | 4.4 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 1.5 | GO:0070198 | protein localization to chromosome, telomeric region(GO:0070198) |
| 0.2 | 1.5 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.2 | 0.4 | GO:0071922 | regulation of sister chromatid cohesion(GO:0007063) regulation of cohesin loading(GO:0071922) |
| 0.2 | 1.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.2 | 0.8 | GO:0099558 | regulation of vesicle fusion(GO:0031338) maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.2 | 0.8 | GO:0072388 | FAD biosynthetic process(GO:0006747) flavin-containing compound metabolic process(GO:0042726) flavin-containing compound biosynthetic process(GO:0042727) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.2 | 1.3 | GO:0031112 | positive regulation of microtubule polymerization or depolymerization(GO:0031112) positive regulation of microtubule polymerization(GO:0031116) |
| 0.2 | 0.8 | GO:0043470 | regulation of glycogen catabolic process(GO:0005981) regulation of carbohydrate catabolic process(GO:0043470) regulation of cellular carbohydrate catabolic process(GO:0043471) |
| 0.2 | 1.2 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 4.5 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.2 | 2.4 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.2 | 0.2 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.2 | 0.8 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.2 | 0.6 | GO:0097237 | cellular response to antibiotic(GO:0071236) cellular response to toxic substance(GO:0097237) |
| 0.2 | 1.0 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 0.2 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.2 | 1.0 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.2 | 1.1 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.2 | 1.5 | GO:0005980 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.2 | 0.7 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.2 | 0.9 | GO:0009794 | regulation of mitotic cell cycle, embryonic(GO:0009794) mitotic cell cycle, embryonic(GO:0045448) |
| 0.2 | 0.7 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.2 | 2.5 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.2 | 1.1 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.2 | 7.7 | GO:0006694 | steroid biosynthetic process(GO:0006694) |
| 0.2 | 0.9 | GO:0071684 | hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.2 | 2.1 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.2 | 1.1 | GO:0019079 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.2 | 0.5 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.2 | 2.1 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 1.7 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 0.9 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.2 | 2.0 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 0.3 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.2 | 0.5 | GO:1902635 | 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate metabolic process(GO:1902633) 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process(GO:1902635) |
| 0.2 | 5.3 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.2 | 0.5 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.2 | 7.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.2 | 0.7 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.2 | 0.7 | GO:1990481 | tRNA pseudouridine synthesis(GO:0031119) mRNA pseudouridine synthesis(GO:1990481) |
| 0.2 | 1.6 | GO:0021634 | optic nerve formation(GO:0021634) |
| 0.2 | 1.0 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 1.5 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.2 | 0.5 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.2 | 0.6 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.2 | 0.5 | GO:2000058 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.2 | 1.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.2 | 0.5 | GO:0030317 | sperm motility(GO:0030317) |
| 0.2 | 2.1 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.2 | 1.1 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.2 | 1.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.2 | 0.5 | GO:0032570 | response to progesterone(GO:0032570) medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.1 | 0.4 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.1 | 1.0 | GO:0060343 | trabecula formation(GO:0060343) heart trabecula formation(GO:0060347) |
| 0.1 | 3.6 | GO:0048920 | posterior lateral line neuromast primordium migration(GO:0048920) |
| 0.1 | 0.6 | GO:0021550 | medulla oblongata development(GO:0021550) dorsal motor nucleus of vagus nerve development(GO:0021744) |
| 0.1 | 4.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.9 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.1 | 0.6 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.1 | 2.7 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 1.0 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 3.8 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.6 | GO:0039015 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.1 | 0.7 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 1.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.7 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.1 | 7.1 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.1 | 6.9 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.1 | 0.4 | GO:0046379 | hyaluronan biosynthetic process(GO:0030213) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.7 | GO:0046292 | formaldehyde metabolic process(GO:0046292) formaldehyde catabolic process(GO:0046294) |
| 0.1 | 0.6 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.1 | 0.4 | GO:0097355 | protein localization to heterochromatin(GO:0097355) |
| 0.1 | 1.6 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 18.0 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.1 | 1.6 | GO:0090559 | regulation of membrane permeability(GO:0090559) |
| 0.1 | 0.4 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.1 | 1.2 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 2.6 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.1 | 2.8 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.1 | 1.1 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.1 | 0.7 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 0.8 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.4 | GO:0043420 | anthranilate metabolic process(GO:0043420) |
| 0.1 | 5.4 | GO:1905037 | autophagosome assembly(GO:0000045) autophagosome organization(GO:1905037) |
| 0.1 | 18.6 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.1 | 1.9 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.4 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 0.5 | GO:0032049 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 3.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.8 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.1 | 0.5 | GO:1990918 | double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.1 | 1.9 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 0.7 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 3.7 | GO:0006360 | transcription from RNA polymerase I promoter(GO:0006360) |
| 0.1 | 1.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 0.6 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 2.4 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 2.0 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 1.0 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.1 | 7.5 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.1 | 0.4 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.1 | 1.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.2 | GO:0044033 | multi-organism metabolic process(GO:0044033) |
| 0.1 | 0.8 | GO:0032196 | transposition(GO:0032196) |
| 0.1 | 0.4 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.1 | 0.9 | GO:0098789 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 1.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.5 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 2.3 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.1 | 0.4 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.1 | 0.2 | GO:0043476 | pigment accumulation(GO:0043476) |
| 0.1 | 0.6 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.1 | 1.7 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.2 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.1 | 0.9 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.1 | 4.7 | GO:0097191 | extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.1 | 1.0 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.1 | 0.8 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 2.3 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.1 | 0.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.9 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.1 | 0.5 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.1 | 7.0 | GO:0031098 | stress-activated protein kinase signaling cascade(GO:0031098) |
| 0.1 | 1.8 | GO:0038061 | NIK/NF-kappaB signaling(GO:0038061) |
| 0.1 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.7 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.1 | 2.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 3.4 | GO:0045055 | regulated exocytosis(GO:0045055) |
| 0.1 | 1.2 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.1 | 0.3 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.1 | 0.7 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.9 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 1.0 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 1.0 | GO:0030202 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.5 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.1 | 2.9 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.1 | 0.2 | GO:0097105 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 1.9 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.1 | 1.0 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.1 | 4.0 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.1 | 0.4 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.1 | 3.4 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.7 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 0.1 | 2.1 | GO:0033014 | porphyrin-containing compound biosynthetic process(GO:0006779) tetrapyrrole biosynthetic process(GO:0033014) |
| 0.1 | 0.7 | GO:0050974 | detection of mechanical stimulus involved in sensory perception(GO:0050974) |
| 0.1 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 1.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.3 | GO:0030214 | hyaluronan catabolic process(GO:0030214) |
| 0.1 | 0.2 | GO:0055016 | hypochord development(GO:0055016) |
| 0.1 | 0.8 | GO:0034204 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.1 | 0.8 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.1 | 1.2 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.1 | 0.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 2.9 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.1 | 0.7 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 0.5 | GO:0019933 | cAMP-mediated signaling(GO:0019933) |
| 0.1 | 2.4 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.1 | 2.1 | GO:1904029 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.1 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 0.5 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.1 | 0.3 | GO:0008334 | histone mRNA metabolic process(GO:0008334) |
| 0.1 | 0.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.9 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.1 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 1.0 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 5.2 | GO:0043547 | positive regulation of GTPase activity(GO:0043547) |
| 0.0 | 0.2 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.2 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.4 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.4 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.1 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.7 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 2.1 | GO:0006887 | exocytosis(GO:0006887) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.3 | GO:0010888 | negative regulation of cholesterol storage(GO:0010887) negative regulation of lipid storage(GO:0010888) |
| 0.0 | 0.2 | GO:0009310 | amine catabolic process(GO:0009310) cellular biogenic amine catabolic process(GO:0042402) |
| 0.0 | 0.1 | GO:0021877 | forebrain neuron fate commitment(GO:0021877) commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.7 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.1 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.9 | GO:0050879 | multicellular organismal movement(GO:0050879) musculoskeletal movement(GO:0050881) |
| 0.0 | 0.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 1.0 | GO:1990542 | mitochondrial transmembrane transport(GO:1990542) |
| 0.0 | 0.9 | GO:0008345 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.3 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.8 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 1.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.6 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 2.7 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 1.3 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 1.0 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.0 | 0.3 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 1.2 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.5 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.0 | 2.8 | GO:0006338 | chromatin remodeling(GO:0006338) |
| 0.0 | 1.8 | GO:0045930 | negative regulation of mitotic cell cycle(GO:0045930) |
| 0.0 | 1.8 | GO:0050953 | sensory perception of light stimulus(GO:0050953) |
| 0.0 | 0.1 | GO:0018342 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 1.4 | GO:0006637 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 0.5 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 2.2 | GO:0006413 | translational initiation(GO:0006413) |
| 0.0 | 0.1 | GO:0052805 | imidazole-containing compound metabolic process(GO:0052803) imidazole-containing compound catabolic process(GO:0052805) |
| 0.0 | 0.4 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.4 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.6 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.2 | GO:0046031 | ADP metabolic process(GO:0046031) |
| 0.0 | 0.2 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.0 | 0.1 | GO:0021982 | pineal gland development(GO:0021982) |
| 0.0 | 0.6 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 1.6 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.2 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.4 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 1.0 | GO:0015698 | inorganic anion transport(GO:0015698) |
| 0.0 | 0.2 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 0.4 | GO:0030217 | T cell differentiation(GO:0030217) |
| 0.0 | 0.1 | GO:1902253 | regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902253) negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.2 | GO:1902622 | regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.5 | GO:0046513 | ceramide biosynthetic process(GO:0046513) |
| 0.0 | 0.1 | GO:0070989 | oxidative demethylation(GO:0070989) |
| 0.0 | 0.3 | GO:0007031 | peroxisome organization(GO:0007031) |
| 0.0 | 0.2 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.3 | GO:0006282 | regulation of DNA repair(GO:0006282) |
| 0.0 | 0.7 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.1 | GO:0072574 | hepatocyte proliferation(GO:0072574) epithelial cell proliferation involved in liver morphogenesis(GO:0072575) |
| 0.0 | 0.0 | GO:1990090 | response to nerve growth factor(GO:1990089) cellular response to nerve growth factor stimulus(GO:1990090) |
| 0.0 | 0.1 | GO:1903670 | regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.9 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.5 | GO:0010951 | negative regulation of endopeptidase activity(GO:0010951) |
| 0.0 | 0.3 | GO:0048232 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 1.1 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 0.7 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 5.8 | GO:0016567 | protein ubiquitination(GO:0016567) |
| 0.0 | 0.2 | GO:0009566 | fertilization(GO:0009566) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 6.9 | GO:0001650 | fibrillar center(GO:0001650) |
| 1.1 | 2.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 1.1 | 7.6 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 1.1 | 5.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 1.0 | 5.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 1.0 | 4.0 | GO:0032019 | mitochondrial cloud(GO:0032019) |
| 1.0 | 4.8 | GO:0070695 | FHF complex(GO:0070695) |
| 0.9 | 6.5 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.8 | 7.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.7 | 2.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.7 | 1.4 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.7 | 2.6 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.6 | 8.0 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.5 | 1.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.5 | 5.6 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.5 | 9.7 | GO:0043186 | P granule(GO:0043186) |
| 0.5 | 2.0 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.5 | 1.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.5 | 3.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.4 | 1.8 | GO:0002144 | cytosolic tRNA wobble base thiouridylase complex(GO:0002144) |
| 0.4 | 3.8 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.4 | 2.4 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.4 | 2.8 | GO:1990131 | EGO complex(GO:0034448) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.4 | 1.9 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.4 | 1.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.4 | 6.7 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.4 | 1.9 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.4 | 1.5 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.4 | 2.2 | GO:0002102 | podosome(GO:0002102) |
| 0.4 | 2.2 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.3 | 1.7 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.3 | 1.0 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.3 | 4.4 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.3 | 18.8 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.3 | 0.3 | GO:0031310 | integral component of vacuolar membrane(GO:0031166) intrinsic component of vacuolar membrane(GO:0031310) |
| 0.3 | 3.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.3 | 3.7 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.3 | 0.9 | GO:0034990 | nuclear cohesin complex(GO:0000798) mitotic cohesin complex(GO:0030892) nuclear mitotic cohesin complex(GO:0034990) nuclear meiotic cohesin complex(GO:0034991) |
| 0.3 | 3.7 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.3 | 0.8 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.3 | 1.1 | GO:0017177 | glucosidase II complex(GO:0017177) |
| 0.3 | 3.5 | GO:0030904 | retromer complex(GO:0030904) |
| 0.3 | 0.8 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.2 | 0.7 | GO:1905202 | methylcrotonoyl-CoA carboxylase complex(GO:1905202) |
| 0.2 | 3.9 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.2 | 1.0 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.2 | 3.8 | GO:0045334 | clathrin-coated endocytic vesicle(GO:0045334) |
| 0.2 | 5.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.2 | 0.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.2 | 0.9 | GO:0031251 | PAN complex(GO:0031251) |
| 0.2 | 2.2 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.2 | 0.6 | GO:1990745 | EARP complex(GO:1990745) |
| 0.2 | 0.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.2 | 0.6 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.2 | 9.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.2 | 1.0 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.2 | 6.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 2.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 2.4 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.2 | 1.6 | GO:0030677 | ribonuclease P complex(GO:0030677) |
| 0.2 | 1.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 3.9 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.2 | 1.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.2 | 22.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.2 | 0.8 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.2 | 1.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.2 | 0.8 | GO:0030062 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.2 | 1.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.2 | 1.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 2.0 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.2 | 0.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.1 | 1.5 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.1 | 0.4 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 0.6 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.1 | 1.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 0.5 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.1 | 1.1 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.6 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 1.1 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.5 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 3.8 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 0.6 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.1 | 1.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 3.1 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.1 | 0.8 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 0.5 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 7.4 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.1 | 1.8 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 2.1 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.1 | 1.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.1 | 1.4 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.1 | 2.5 | GO:0030496 | midbody(GO:0030496) |
| 0.1 | 4.0 | GO:0031970 | mitochondrial intermembrane space(GO:0005758) organelle envelope lumen(GO:0031970) |
| 0.1 | 4.6 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 7.1 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.1 | 0.3 | GO:0031417 | NatC complex(GO:0031417) |
| 0.1 | 1.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 8.9 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 1.1 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.1 | 12.9 | GO:0005813 | centrosome(GO:0005813) |
| 0.1 | 1.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 0.4 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 2.9 | GO:0016529 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 0.6 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.1 | 0.8 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 0.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 0.7 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.9 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.2 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.1 | 0.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 0.9 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 4.4 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 1.2 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.7 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 3.3 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.1 | 2.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 1.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 2.0 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.1 | 1.0 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.1 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 2.4 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.1 | 2.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.7 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 4.7 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 1.5 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 4.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 1.4 | GO:0030133 | transport vesicle(GO:0030133) |
| 0.0 | 0.8 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.9 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.1 | GO:0071159 | NF-kappaB complex(GO:0071159) |
| 0.0 | 0.2 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.6 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 1.7 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 2.3 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 1.6 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:1902555 | endoribonuclease complex(GO:1902555) |
| 0.0 | 1.6 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 2.4 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 0.4 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 1.0 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 12.2 | GO:0005794 | Golgi apparatus(GO:0005794) |
| 0.0 | 0.5 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 1.9 | GO:0000228 | nuclear chromosome(GO:0000228) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 8.7 | GO:0005739 | mitochondrion(GO:0005739) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 10.9 | GO:0004937 | alpha-adrenergic receptor activity(GO:0004936) alpha1-adrenergic receptor activity(GO:0004937) |
| 2.0 | 6.1 | GO:0030629 | U6 snRNA 3'-end binding(GO:0030629) 23S rRNA (adenine(1618)-N(6))-methyltransferase activity(GO:0052907) |
| 1.6 | 9.8 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 1.3 | 5.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 1.2 | 4.8 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 1.1 | 17.9 | GO:0032190 | acrosin binding(GO:0032190) |
| 1.0 | 4.2 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 1.0 | 11.0 | GO:0008494 | translation activator activity(GO:0008494) |
| 1.0 | 7.8 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.9 | 2.7 | GO:0034417 | bisphosphoglycerate 3-phosphatase activity(GO:0034417) |
| 0.8 | 4.2 | GO:0052833 | inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.8 | 2.5 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.8 | 2.5 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.8 | 14.6 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.7 | 19.7 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.7 | 2.9 | GO:0004061 | arylformamidase activity(GO:0004061) |
| 0.7 | 2.2 | GO:0001635 | adrenomedullin receptor activity(GO:0001605) calcitonin gene-related peptide receptor activity(GO:0001635) calcitonin receptor activity(GO:0004948) calcitonin family receptor activity(GO:0097642) |
| 0.7 | 13.6 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.7 | 2.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) cGMP binding(GO:0030553) |
| 0.7 | 2.7 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.7 | 2.0 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.7 | 2.7 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.7 | 4.0 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.7 | 4.0 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.7 | 3.9 | GO:0034338 | short-chain carboxylesterase activity(GO:0034338) |
| 0.6 | 2.6 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.6 | 1.9 | GO:0031729 | CCR1 chemokine receptor binding(GO:0031726) CCR4 chemokine receptor binding(GO:0031729) CCR5 chemokine receptor binding(GO:0031730) |
| 0.6 | 1.9 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.6 | 1.8 | GO:0016843 | amine-lyase activity(GO:0016843) strictosidine synthase activity(GO:0016844) |
| 0.6 | 5.9 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.6 | 2.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.6 | 2.3 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.6 | 2.3 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.6 | 7.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.5 | 2.1 | GO:0072572 | poly-ADP-D-ribose binding(GO:0072572) |
| 0.5 | 2.1 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.5 | 2.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.5 | 14.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.5 | 1.5 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.5 | 3.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.5 | 1.5 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.5 | 1.5 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.5 | 5.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.5 | 5.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.4 | 1.3 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.4 | 1.8 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.4 | 4.4 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.4 | 1.3 | GO:0046592 | polyamine oxidase activity(GO:0046592) |
| 0.4 | 1.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.4 | 1.3 | GO:0004170 | dUTP diphosphatase activity(GO:0004170) |
| 0.4 | 1.6 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.4 | 4.4 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.4 | 1.6 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.4 | 1.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.4 | 3.6 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.4 | 9.2 | GO:0032813 | tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.4 | 1.9 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.4 | 1.1 | GO:0004377 | GDP-Man:Man3GlcNAc2-PP-Dol alpha-1,2-mannosyltransferase activity(GO:0004377) |
| 0.4 | 5.2 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.4 | 1.5 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.4 | 2.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.4 | 1.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.4 | 3.9 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.4 | 3.2 | GO:0017091 | AU-rich element binding(GO:0017091) mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.3 | 1.7 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.3 | 19.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.3 | 1.4 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.3 | 1.0 | GO:0047464 | heparosan-N-sulfate-glucuronate 5-epimerase activity(GO:0047464) |
| 0.3 | 0.3 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.3 | 1.9 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.3 | 5.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.3 | 2.4 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.3 | 1.5 | GO:0008184 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.3 | 0.9 | GO:0070513 | death domain binding(GO:0070513) |
| 0.3 | 4.1 | GO:0015368 | calcium:cation antiporter activity(GO:0015368) |
| 0.3 | 1.2 | GO:0005460 | UDP-glucose transmembrane transporter activity(GO:0005460) |
| 0.3 | 1.2 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.3 | 1.9 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.3 | 1.4 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.3 | 0.8 | GO:0035514 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.3 | 2.7 | GO:0031995 | insulin-like growth factor I binding(GO:0031994) insulin-like growth factor II binding(GO:0031995) |
| 0.3 | 1.1 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 0.3 | 1.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.3 | 2.1 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.3 | 2.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 1.5 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.2 | 3.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.2 | 1.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 1.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.2 | 3.9 | GO:0052744 | phosphatidylinositol monophosphate phosphatase activity(GO:0052744) |
| 0.2 | 5.8 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.2 | 2.9 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 2.6 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.2 | 0.7 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.2 | 2.1 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.2 | 1.2 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 3.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 1.4 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.2 | 0.7 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 1.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.2 | 2.0 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.2 | 2.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.2 | 1.1 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.2 | 1.3 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 0.7 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.2 | 0.9 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.2 | 0.8 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.2 | 0.8 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.2 | 1.3 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.2 | 1.0 | GO:1990756 | protein binding, bridging involved in substrate recognition for ubiquitination(GO:1990756) |
| 0.2 | 3.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.2 | 4.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 1.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.2 | 0.6 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 1.8 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 8.4 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.2 | 0.6 | GO:0033745 | L-methionine-(R)-S-oxide reductase activity(GO:0033745) |
| 0.2 | 0.6 | GO:0016595 | glutamate-cysteine ligase activity(GO:0004357) glutamate binding(GO:0016595) |
| 0.2 | 4.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.2 | 0.6 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.2 | 0.2 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.2 | 1.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.2 | 1.3 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.2 | 0.7 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.2 | 2.0 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.2 | 3.3 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.2 | 11.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.2 | 1.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 0.5 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.2 | 1.5 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 0.5 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 1.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.2 | 1.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 2.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.2 | 0.7 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 4.2 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.2 | 0.8 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.2 | 5.5 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.2 | 1.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.2 | 3.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.2 | 2.0 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.2 | 0.8 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.2 | 0.6 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 1.0 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.1 | 0.6 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.1 | 3.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 0.8 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 0.8 | GO:0099589 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin receptor activity(GO:0099589) |
| 0.1 | 0.4 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 1.2 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 0.4 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.1 | 0.5 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.1 | 0.4 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.4 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.1 | 0.9 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.1 | 2.4 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 1.9 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.1 | 3.0 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.1 | 0.4 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.1 | 0.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 3.7 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 1.6 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 1.9 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 3.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 1.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.1 | 0.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.7 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.8 | GO:0070740 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.4 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.1 | 0.7 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.5 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.1 | 0.4 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.1 | 1.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.6 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.8 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 1.4 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.7 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 0.7 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 4.1 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.4 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.1 | 22.6 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.1 | 2.6 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.1 | 0.5 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 1.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 1.6 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 0.7 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.1 | 0.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 1.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 11.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 0.9 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.8 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 2.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.4 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 1.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 4.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.1 | 1.8 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 1.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.9 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.1 | 2.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.1 | 0.3 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.1 | 0.9 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.7 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 0.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.1 | 6.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.1 | 3.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.2 | GO:0052717 | tRNA-specific adenosine-34 deaminase activity(GO:0052717) |
| 0.1 | 0.3 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.1 | 0.3 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.1 | 0.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.5 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.9 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 1.6 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.1 | 1.1 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.1 | 4.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 0.2 | GO:0043121 | neurotrophin binding(GO:0043121) nerve growth factor binding(GO:0048406) |
| 0.1 | 0.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.8 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.4 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.9 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 2.7 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.3 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.7 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.7 | GO:0015245 | long-chain fatty acid transporter activity(GO:0005324) fatty acid transporter activity(GO:0015245) |
| 0.0 | 1.7 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 1.1 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.7 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 5.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 1.5 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.0 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.4 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.4 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.7 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.0 | 0.5 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 5.1 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 1.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 1.8 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 2.3 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 1.1 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 11.0 | GO:0004674 | protein serine/threonine kinase activity(GO:0004674) |
| 0.0 | 3.9 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.4 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.6 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 5.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.3 | GO:0015296 | anion:cation symporter activity(GO:0015296) |
| 0.0 | 0.3 | GO:0022884 | protein transmembrane transporter activity(GO:0008320) macromolecule transmembrane transporter activity(GO:0022884) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.0 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.9 | GO:0031072 | heat shock protein binding(GO:0031072) |
| 0.0 | 0.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.3 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.1 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.7 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 2.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.3 | 5.9 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 0.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 5.7 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.2 | 4.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.2 | 3.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.2 | 3.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.2 | 3.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 3.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.2 | 1.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 0.4 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.1 | 1.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 1.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 0.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 1.9 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.1 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 0.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 1.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 1.9 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 1.1 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 2.8 | PID FOXO PATHWAY | FoxO family signaling |
| 0.1 | 3.5 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 3.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 0.9 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 0.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 0.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 0.8 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.1 | 0.8 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.0 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.2 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.0 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.2 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.8 | 2.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.7 | 2.2 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.6 | 5.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.6 | 1.9 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.5 | 3.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.4 | 2.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.4 | 1.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.4 | 2.3 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.4 | 6.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.3 | 2.6 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.3 | 5.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.3 | 1.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.3 | 3.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.2 | 1.7 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.2 | 1.7 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.2 | 1.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 4.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 2.3 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.2 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.2 | 1.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.2 | 5.8 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.2 | 3.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.2 | 2.7 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.2 | 1.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.2 | 4.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.2 | 2.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.2 | 1.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.2 | 0.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.1 | 2.3 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 2.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 1.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 1.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 2.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 0.6 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 1.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.8 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 4.2 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 0.8 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 1.0 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 5.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.1 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 4.0 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 1.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 3.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.2 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 0.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 0.6 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.1 | 0.6 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.1 | 1.2 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 0.5 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.1 | 1.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.1 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 0.7 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 1.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 0.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.1 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 0.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 3.9 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.1 | 0.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.3 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 0.5 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 1.2 | REACTOME TELOMERE MAINTENANCE | Genes involved in Telomere Maintenance |
| 0.0 | 0.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.3 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.9 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 3.7 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.1 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 0.6 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |