Project
DANIO-CODE
Navigation
Downloads
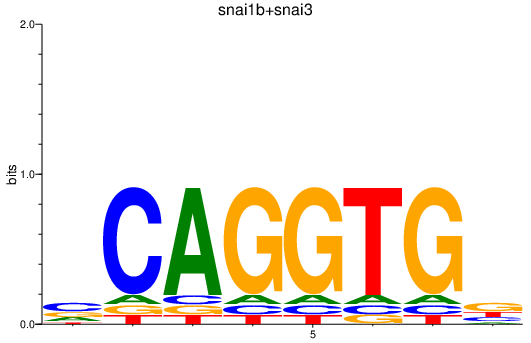
Results for snai1b+snai3
Z-value: 0.81
Transcription factors associated with snai1b+snai3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
snai3
|
ENSDARG00000031243 | snail family zinc finger 3 |
|
snai1b
|
ENSDARG00000046019 | snail family zinc finger 1b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| snai1b | dr10_dc_chr23_-_2573949_2573956 | -0.61 | 1.2e-02 | Click! |
| snai3 | dr10_dc_chr7_+_54811485_54811499 | 0.54 | 2.9e-02 | Click! |
Activity profile of snai1b+snai3 motif
Sorted Z-values of snai1b+snai3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of snai1b+snai3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_58448953 | 3.60 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr1_+_41762057 | 3.25 |
ENSDART00000137609
|
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr7_-_55346967 | 2.70 |
ENSDART00000135304
|
pabpn1l
|
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
| chr22_+_25113293 | 2.08 |
ENSDART00000171851
|
si:ch211-226h8.4
|
si:ch211-226h8.4 |
| chr9_-_28444272 | 1.79 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr13_-_21541531 | 1.77 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr12_+_22551105 | 1.69 |
ENSDART00000123808
ENSDART00000159864 |
cdca9
|
cell division cycle associated 9 |
| chr20_-_23527234 | 1.66 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr7_+_58449163 | 1.66 |
ENSDART00000167166
|
zgc:56231
|
zgc:56231 |
| chr19_+_33552393 | 1.57 |
ENSDART00000043039
|
fam84b
|
family with sequence similarity 84, member B |
| chr10_+_37229202 | 1.53 |
ENSDART00000136510
|
ksr1a
|
kinase suppressor of ras 1a |
| chr12_-_2834771 | 1.52 |
ENSDART00000114854
ENSDART00000163759 |
zdhhc16b
|
zinc finger, DHHC-type containing 16b |
| chr9_-_28444024 | 1.46 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr5_-_11530803 | 1.42 |
ENSDART00000159896
|
gatsl3
|
GATS protein-like 3 |
| chr7_+_58449127 | 1.40 |
ENSDART00000167166
|
zgc:56231
|
zgc:56231 |
| chr22_+_22413803 | 1.36 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr20_-_43846604 | 1.17 |
ENSDART00000150078
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr5_-_23192997 | 1.09 |
ENSDART00000167629
|
rnf128a
|
ring finger protein 128a |
| chr12_-_35467285 | 1.03 |
ENSDART00000160336
|
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr22_-_26231695 | 0.93 |
ENSDART00000142821
|
ccdc130
|
coiled-coil domain containing 130 |
| chr9_-_28443972 | 0.93 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr22_+_2237813 | 0.90 |
|
|
|
| chr5_-_23192934 | 0.87 |
ENSDART00000021462
|
rnf128a
|
ring finger protein 128a |
| chr7_+_58448909 | 0.85 |
ENSDART00000024185
|
zgc:56231
|
zgc:56231 |
| chr8_-_10912137 | 0.76 |
ENSDART00000064058
ENSDART00000162109 ENSDART00000101277 |
pqlc2
|
PQ loop repeat containing 2 |
| chr20_-_43846553 | 0.75 |
ENSDART00000150078
|
si:dkeyp-50f7.2
|
si:dkeyp-50f7.2 |
| chr19_-_32314264 | 0.72 |
|
|
|
| chr25_+_27301039 | 0.70 |
ENSDART00000149456
|
wasla
|
Wiskott-Aldrich syndrome-like a |
| chr6_-_10492724 | 0.68 |
ENSDART00000002247
|
sp3b
|
Sp3b transcription factor |
| chr16_-_29470491 | 0.67 |
ENSDART00000175571
|
tlr18
|
toll-like receptor 18 |
| chr12_+_16831493 | 0.64 |
ENSDART00000020824
|
pank1b
|
pantothenate kinase 1b |
| chr23_+_34120843 | 0.63 |
ENSDART00000143339
|
chchd6a
|
coiled-coil-helix-coiled-coil-helix domain containing 6a |
| chr22_+_22413878 | 0.62 |
ENSDART00000147825
|
kif14
|
kinesin family member 14 |
| chr2_-_56632969 | 0.61 |
ENSDART00000089158
|
hmha1a
|
histocompatibility (minor) HA-1 a |
| chr21_-_35291005 | 0.61 |
ENSDART00000134780
|
ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1 |
| chr15_+_1238560 | 0.60 |
ENSDART00000167760
ENSDART00000163827 |
mfsd1
|
major facilitator superfamily domain containing 1 |
| chr15_+_12127861 | 0.58 |
|
|
|
| chr9_-_28444210 | 0.58 |
ENSDART00000160387
|
ccnyl1
|
cyclin Y-like 1 |
| chr1_+_40134375 | 0.57 |
ENSDART00000177517
|
CR387992.1
|
ENSDARG00000108790 |
| chr7_+_73583860 | 0.57 |
ENSDART00000166244
|
si:ch73-252p3.1
|
si:ch73-252p3.1 |
| chr6_-_37771729 | 0.56 |
ENSDART00000078324
|
nipa1
|
non imprinted in Prader-Willi/Angelman syndrome 1 |
| chr16_+_41110027 | 0.55 |
ENSDART00000141130
|
scap
|
SREBF chaperone |
| chr7_+_67475893 | 0.55 |
ENSDART00000160086
|
zgc:162592
|
zgc:162592 |
| chr14_-_30747372 | 0.55 |
ENSDART00000002250
|
hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr1_-_529071 | 0.55 |
ENSDART00000147610
|
si:ch73-41e3.7
|
si:ch73-41e3.7 |
| chr23_+_34121048 | 0.50 |
ENSDART00000143339
|
chchd6a
|
coiled-coil-helix-coiled-coil-helix domain containing 6a |
| chr22_-_26231606 | 0.49 |
ENSDART00000060888
|
ccdc130
|
coiled-coil domain containing 130 |
| chr17_+_24571678 | 0.46 |
ENSDART00000092941
|
rlf
|
rearranged L-myc fusion |
| chr7_+_20829933 | 0.41 |
ENSDART00000173612
|
serpinh2
|
serine (or cysteine) peptidase inhibitor, clade H, member 2 |
| chr5_-_68712851 | 0.41 |
ENSDART00000109487
|
CABZ01032476.1
|
ENSDARG00000074645 |
| chr20_-_25731057 | 0.36 |
|
|
|
| KN150232v1_+_13079 | 0.36 |
ENSDART00000174701
|
CABZ01084889.1
|
ENSDARG00000107269 |
| chr3_+_14492079 | 0.34 |
ENSDART00000091070
|
zgc:158403
|
zgc:158403 |
| chr11_+_6893104 | 0.33 |
ENSDART00000144006
|
crtc1b
|
CREB regulated transcription coactivator 1b |
| chr8_-_17736169 | 0.33 |
ENSDART00000063592
ENSDART00000175064 |
prkcz
|
protein kinase C, zeta |
| chr25_+_27300835 | 0.33 |
ENSDART00000103519
|
wasla
|
Wiskott-Aldrich syndrome-like a |
| chr24_-_41160119 | 0.31 |
ENSDART00000173529
ENSDART00000159738 |
ttc21a
|
tetratricopeptide repeat domain 21A |
| chr5_-_21548930 | 0.30 |
ENSDART00000002938
|
mtmr8
|
myotubularin related protein 8 |
| chr6_-_21756764 | 0.28 |
ENSDART00000167937
|
p4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr13_-_21541412 | 0.25 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr25_+_5232982 | 0.24 |
|
|
|
| chr14_-_14849759 | 0.22 |
ENSDART00000172666
|
uvssa
|
UV-stimulated scaffold protein A |
| chr2_-_56632913 | 0.22 |
ENSDART00000089158
|
hmha1a
|
histocompatibility (minor) HA-1 a |
| chr7_-_30654776 | 0.20 |
ENSDART00000075421
|
sord
|
sorbitol dehydrogenase |
| chr13_-_36400356 | 0.19 |
ENSDART00000175311
|
pacs2
|
phosphofurin acidic cluster sorting protein 2 |
| chr5_-_11530880 | 0.18 |
ENSDART00000159896
|
gatsl3
|
GATS protein-like 3 |
| chr22_-_26504565 | 0.18 |
ENSDART00000087623
|
zgc:194330
|
zgc:194330 |
| chr17_-_25865861 | 0.17 |
|
|
|
| chr16_-_9939497 | 0.16 |
ENSDART00000148528
|
grhl2a
|
grainyhead-like transcription factor 2a |
| chr21_-_26678542 | 0.14 |
ENSDART00000053794
|
banf1
|
barrier to autointegration factor 1 |
| chr5_-_68712571 | 0.14 |
ENSDART00000109487
|
CABZ01032476.1
|
ENSDARG00000074645 |
| chr23_-_3813451 | 0.13 |
|
|
|
| chr23_-_29897606 | 0.12 |
ENSDART00000020616
|
zgc:194189
|
zgc:194189 |
| chr23_+_42449089 | 0.10 |
ENSDART00000171119
|
cyp2aa9
|
cytochrome P450, family 2, subfamily AA, polypeptide 9 |
| chr20_-_21773327 | 0.08 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr8_+_21974863 | 0.07 |
|
|
|
| chr23_+_42779461 | 0.07 |
|
|
|
| chr14_-_30747259 | 0.06 |
ENSDART00000002250
|
hs6st2
|
heparan sulfate 6-O-sulfotransferase 2 |
| chr9_+_331815 | 0.06 |
ENSDART00000170312
|
actr5
|
ARP5 actin related protein 5 homolog |
| chr20_-_23527004 | 0.05 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr16_-_22394481 | 0.03 |
|
|
|
| chr4_-_12792010 | 0.03 |
ENSDART00000067131
|
irak3
|
interleukin-1 receptor-associated kinase 3 |
| chr25_+_5233045 | 0.01 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.7 | 4.8 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.2 | 0.5 | GO:0031048 | chromatin silencing by small RNA(GO:0031048) |
| 0.1 | 1.0 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 1.0 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 2.0 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 1.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.5 | GO:0071880 | adenylate cyclase-activating adrenergic receptor signaling pathway(GO:0071880) |
| 0.1 | 0.3 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 9.5 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.7 | GO:0002224 | toll-like receptor signaling pathway(GO:0002224) |
| 0.0 | 0.3 | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) |
| 0.0 | 0.2 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.0 | 1.5 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0019405 | alditol catabolic process(GO:0019405) |
| 0.0 | 0.2 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 1.5 | GO:0000070 | mitotic sister chromatid segregation(GO:0000070) |
| 0.0 | 1.4 | GO:0017148 | negative regulation of translation(GO:0017148) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.2 | 9.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 1.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 1.0 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 3.2 | GO:0030424 | axon(GO:0030424) |
| 0.0 | 2.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 2.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.0 | GO:0005884 | actin filament(GO:0005884) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:1903231 | mRNA binding involved in posttranscriptional gene silencing(GO:1903231) |
| 0.2 | 9.5 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.1 | 0.5 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.6 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 2.0 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 4.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 0.5 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.1 | 3.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 1.5 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.3 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 2.1 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.0 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |