Project
DANIO-CODE
Navigation
Downloads
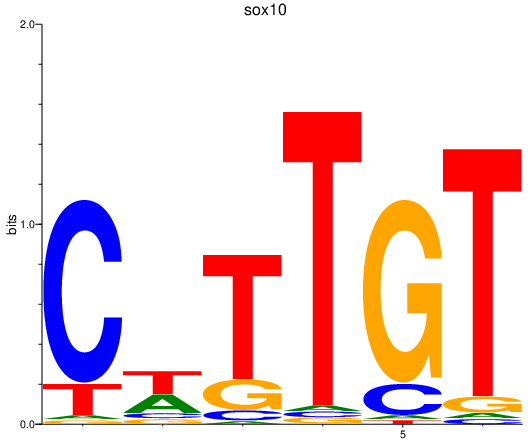
Results for sox10
Z-value: 1.31
Transcription factors associated with sox10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox10
|
ENSDARG00000077467 | SRY-box transcription factor 10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox10 | dr10_dc_chr3_+_2013046_2013122 | 0.19 | 4.8e-01 | Click! |
Activity profile of sox10 motif
Sorted Z-values of sox10 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of sox10
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_45155460 | 5.05 |
ENSDART00000170767
|
ved
|
ventrally expressed dharma/bozozok antagonist |
| chr1_-_50066633 | 3.34 |
|
|
|
| chr5_+_26925238 | 2.68 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr5_-_24982964 | 2.67 |
ENSDART00000031665
ENSDART00000161043 |
anxa1a
|
annexin A1a |
| chr7_+_24770873 | 2.43 |
ENSDART00000165314
|
AL954715.1
|
ENSDARG00000103682 |
| chr13_+_22545318 | 2.37 |
ENSDART00000143312
|
zgc:193505
|
zgc:193505 |
| chr9_-_18903879 | 2.35 |
ENSDART00000138785
|
si:dkey-239h2.3
|
si:dkey-239h2.3 |
| chr16_-_51543374 | 2.13 |
ENSDART00000090530
|
si:ch211-107n13.1
|
si:ch211-107n13.1 |
| chr1_-_50831155 | 1.93 |
ENSDART00000152719
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr6_-_39315024 | 1.89 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr3_+_24067387 | 1.85 |
ENSDART00000055609
|
atf4b
|
activating transcription factor 4b |
| chr16_+_4825019 | 1.78 |
ENSDART00000167665
|
LIN28A
|
lin-28 homolog A |
| chr8_-_38168395 | 1.70 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr15_-_1625846 | 1.69 |
ENSDART00000081875
|
nnr
|
nanor |
| chr18_+_40365208 | 1.66 |
ENSDART00000167134
|
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr25_-_34616997 | 1.63 |
ENSDART00000137502
|
si:ch73-36p18.1
|
si:ch73-36p18.1 |
| chr1_-_31123999 | 1.61 |
ENSDART00000152493
|
nt5c2b
|
5'-nucleotidase, cytosolic IIb |
| chr1_-_18695214 | 1.56 |
|
|
|
| chr23_-_24468191 | 1.51 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr1_+_16683931 | 1.50 |
ENSDART00000103262
ENSDART00000145068 ENSDART00000169619 ENSDART00000010526 |
fat1a
|
FAT atypical cadherin 1a |
| chr23_+_24711233 | 1.50 |
|
|
|
| chr17_-_43896998 | 1.43 |
ENSDART00000156513
|
BX465227.1
|
ENSDARG00000097825 |
| chr13_-_33137075 | 1.39 |
ENSDART00000138096
|
oip5-as1
|
oip5 antisense RNA 1 |
| chr8_-_50992034 | 1.39 |
ENSDART00000004065
|
zgc:91909
|
zgc:91909 |
| chr7_+_56076854 | 1.38 |
|
|
|
| chr4_-_11604184 | 1.38 |
ENSDART00000170136
|
net1
|
neuroepithelial cell transforming 1 |
| chr20_-_21773202 | 1.35 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr22_+_17091411 | 1.31 |
ENSDART00000170076
|
frem1b
|
Fras1 related extracellular matrix 1b |
| chr8_-_16662185 | 1.30 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr14_-_30556788 | 1.29 |
ENSDART00000087918
|
slc7a3b
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3b |
| chr3_-_16100715 | 1.28 |
ENSDART00000146699
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr13_-_46292762 | 1.28 |
ENSDART00000149125
|
fgfr2
|
fibroblast growth factor receptor 2 |
| chr18_-_5796081 | 1.26 |
ENSDART00000097696
ENSDART00000151453 |
si:ch1073-459b3.2
|
si:ch1073-459b3.2 |
| chr22_-_36561247 | 1.26 |
ENSDART00000056188
|
polr2h
|
info polymerase (RNA) II (DNA directed) polypeptide H |
| chr4_-_7721419 | 1.22 |
|
|
|
| chr21_-_44610563 | 1.20 |
ENSDART00000099528
|
spry4
|
sprouty homolog 4 (Drosophila) |
| chr1_+_27174549 | 1.20 |
ENSDART00000102337
|
dnajb14
|
DnaJ (Hsp40) homolog, subfamily B, member 14 |
| KN149859v1_+_16751 | 1.13 |
ENSDART00000167610
|
ENSDARG00000103399
|
ENSDARG00000103399 |
| chr3_+_30962542 | 1.12 |
ENSDART00000153074
|
si:dkey-66i24.9
|
si:dkey-66i24.9 |
| chr25_-_12691849 | 1.12 |
ENSDART00000158551
|
slc7a5
|
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
| chr17_-_48722470 | 1.12 |
ENSDART00000160893
|
kif6
|
kinesin family member 6 |
| chr6_-_43094573 | 1.12 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr22_+_14073249 | 1.11 |
ENSDART00000080335
ENSDART00000018432 |
he1b
|
hatching enzyme 1b |
| chr14_-_31184501 | 1.09 |
|
|
|
| chr14_-_7774262 | 1.09 |
ENSDART00000045109
|
zgc:92242
|
zgc:92242 |
| chr20_-_21773327 | 1.08 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr2_-_22692538 | 1.07 |
|
|
|
| chr17_+_24830545 | 1.05 |
ENSDART00000062917
|
cx35.4
|
connexin 35.4 |
| chr13_-_13163676 | 1.03 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr14_+_38505346 | 1.02 |
ENSDART00000129958
ENSDART00000144599 |
hnrnpa0b
|
heterogeneous nuclear ribonucleoprotein A0b |
| chr5_+_56885728 | 1.02 |
|
|
|
| chr18_-_782643 | 1.01 |
ENSDART00000161084
|
si:dkey-205h23.1
|
si:dkey-205h23.1 |
| chr25_+_31457309 | 0.94 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr5_-_14148028 | 0.93 |
ENSDART00000113037
|
sema4c
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr3_-_26073676 | 0.92 |
ENSDART00000169123
|
gdpd3a
|
glycerophosphodiester phosphodiesterase domain containing 3a |
| chr3_-_38642067 | 0.92 |
ENSDART00000155518
|
znf281a
|
zinc finger protein 281a |
| chr23_+_18565721 | 0.91 |
|
|
|
| chr13_+_1051015 | 0.88 |
ENSDART00000033528
|
tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr8_-_52729505 | 0.87 |
ENSDART00000168241
|
tubb2b
|
tubulin, beta 2b |
| chr9_+_36023553 | 0.86 |
ENSDART00000122169
|
si:dkey-67c22.2
|
si:dkey-67c22.2 |
| chr22_-_15567180 | 0.86 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr1_-_44009572 | 0.85 |
ENSDART00000144900
|
ENSDARG00000079632
|
ENSDARG00000079632 |
| chr20_+_29307142 | 0.81 |
ENSDART00000153016
|
katnbl1
|
katanin p80 subunit B-like 1 |
| chr22_-_3661536 | 0.81 |
ENSDART00000153634
|
FP565432.1
|
ENSDARG00000097145 |
| chr12_-_30662709 | 0.81 |
ENSDART00000126466
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr5_-_43686611 | 0.81 |
ENSDART00000146080
|
si:ch73-337l15.2
|
si:ch73-337l15.2 |
| chr19_-_3208938 | 0.80 |
|
|
|
| chr19_+_31817286 | 0.79 |
ENSDART00000078459
|
tmem55a
|
transmembrane protein 55A |
| chr8_+_2517472 | 0.79 |
ENSDART00000112914
|
si:ch211-51h9.7
|
si:ch211-51h9.7 |
| chr7_-_2367001 | 0.78 |
ENSDART00000174011
|
si:cabz01007812.1
|
si:cabz01007812.1 |
| chr11_+_6138968 | 0.76 |
ENSDART00000082223
|
tax1bp3
|
Tax1 (human T-cell leukemia virus type I) binding protein 3 |
| chr21_-_25719352 | 0.74 |
ENSDART00000002341
|
cldnc
|
claudin c |
| chr2_-_43110857 | 0.74 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr14_+_35880163 | 0.73 |
ENSDART00000148319
|
pitx2
|
paired-like homeodomain 2 |
| chr8_+_23334921 | 0.73 |
ENSDART00000085361
ENSDART00000046460 ENSDART00000145062 |
dnmt3ba
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate a |
| chr20_-_22576513 | 0.73 |
ENSDART00000103510
|
pdgfra
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr12_+_31623806 | 0.73 |
|
|
|
| chr13_-_46292975 | 0.72 |
ENSDART00000167402
ENSDART00000080914 ENSDART00000080916 |
fgfr2
|
fibroblast growth factor receptor 2 |
| chr6_-_27963880 | 0.72 |
ENSDART00000163747
|
BX914220.1
|
ENSDARG00000098603 |
| chr10_+_45138195 | 0.72 |
ENSDART00000162566
|
h2afvb
|
H2A histone family, member Vb |
| chr1_-_50576110 | 0.71 |
ENSDART00000146612
|
esf1
|
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) |
| chr16_+_40610589 | 0.71 |
ENSDART00000038294
|
tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr4_+_50923792 | 0.70 |
ENSDART00000161370
|
BX640547.3
|
ENSDARG00000100076 |
| chr3_-_58590651 | 0.70 |
ENSDART00000057640
|
dhrs7ca
|
dehydrogenase/reductase (SDR family) member 7Ca |
| chr6_-_49512935 | 0.69 |
ENSDART00000166238
|
rplp2
|
ribosomal protein, large P2 |
| chr3_+_30962827 | 0.68 |
ENSDART00000153074
|
si:dkey-66i24.9
|
si:dkey-66i24.9 |
| chr17_+_1986151 | 0.67 |
ENSDART00000103775
|
pak6a
|
p21 protein (Cdc42/Rac)-activated kinase 6a |
| chr9_-_34491458 | 0.65 |
ENSDART00000049805
|
ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr17_+_24045786 | 0.65 |
ENSDART00000132755
|
b3gnt2b
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2b |
| chr13_-_13163801 | 0.65 |
ENSDART00000156968
|
fgfr3
|
fibroblast growth factor receptor 3 |
| chr6_+_10491884 | 0.64 |
ENSDART00000151017
|
BX936397.2
|
ENSDARG00000096373 |
| chr7_-_6281491 | 0.64 |
ENSDART00000172795
|
si:ch73-368j24.12
|
si:ch73-368j24.12 |
| chr21_-_39900518 | 0.63 |
ENSDART00000175055
|
CABZ01065291.1
|
ENSDARG00000107374 |
| chr15_-_17818432 | 0.63 |
ENSDART00000157198
|
CT573342.1
|
ENSDARG00000097333 |
| chr16_+_53238110 | 0.59 |
ENSDART00000102170
|
CABZ01053976.1
|
ENSDARG00000069929 |
| chr16_-_26858511 | 0.58 |
ENSDART00000135860
|
rnf41l
|
ring finger protein 41, like |
| chr6_-_43094926 | 0.57 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr11_+_37345024 | 0.57 |
ENSDART00000077496
|
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr9_+_36023025 | 0.55 |
ENSDART00000122169
|
si:dkey-67c22.2
|
si:dkey-67c22.2 |
| chr10_-_8336541 | 0.55 |
ENSDART00000163803
|
plpp1a
|
phospholipid phosphatase 1a |
| chr11_+_30048913 | 0.54 |
|
|
|
| chr7_+_20008453 | 0.53 |
ENSDART00000012450
|
dvl2
|
dishevelled segment polarity protein 2 |
| chr18_+_45564871 | 0.53 |
|
|
|
| chr12_+_18622682 | 0.53 |
ENSDART00000153456
|
mkl1b
|
megakaryoblastic leukemia (translocation) 1b |
| chr20_-_25726868 | 0.53 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr25_+_20618512 | 0.51 |
ENSDART00000144748
|
ergic2
|
ERGIC and golgi 2 |
| chr11_+_30049041 | 0.50 |
|
|
|
| chr5_+_25521198 | 0.50 |
ENSDART00000002221
|
bco2l
|
beta-carotene 15, 15-dioxygenase 2, like |
| chr7_+_49391403 | 0.50 |
ENSDART00000131210
|
rassf7b
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7b |
| chr22_-_20284307 | 0.49 |
ENSDART00000159604
|
mbd3b
|
methyl-CpG binding domain protein 3b |
| chr11_+_25063488 | 0.48 |
|
|
|
| chr1_-_23603046 | 0.48 |
|
|
|
| chr9_+_38975508 | 0.48 |
|
|
|
| chr7_-_71092394 | 0.48 |
ENSDART00000081245
|
dhx15
|
DEAH (Asp-Glu-Ala-His) box helicase 15 |
| chr7_+_19583578 | 0.47 |
ENSDART00000149812
|
ovol1
|
ovo-like zinc finger 1 |
| chr5_+_39619826 | 0.47 |
ENSDART00000114078
|
si:dkey-193c22.2
|
si:dkey-193c22.2 |
| chr14_+_38509121 | 0.46 |
ENSDART00000052511
|
hnrnpa0l
|
heterogeneous nuclear ribonucleoprotein A0, like |
| chr2_-_42510909 | 0.46 |
ENSDART00000144274
|
si:dkey-7l6.3
|
si:dkey-7l6.3 |
| chr5_-_51351827 | 0.45 |
ENSDART00000097194
|
serinc5
|
serine incorporator 5 |
| chr20_-_47123598 | 0.45 |
ENSDART00000100320
|
dnmt3aa
|
DNA (cytosine-5-)-methyltransferase 3 alpha a |
| chr8_-_12171624 | 0.45 |
ENSDART00000132824
|
dab2ipa
|
DAB2 interacting protein a |
| chr13_-_46292862 | 0.45 |
ENSDART00000150061
|
fgfr2
|
fibroblast growth factor receptor 2 |
| chr22_-_18521227 | 0.45 |
ENSDART00000105404
|
cirbpb
|
cold inducible RNA binding protein b |
| chr21_-_25704662 | 0.44 |
ENSDART00000101211
|
cldnh
|
claudin h |
| chr17_-_6191539 | 0.44 |
ENSDART00000081707
|
ENSDARG00000058775
|
ENSDARG00000058775 |
| chr15_-_16140187 | 0.43 |
ENSDART00000080413
|
srsf1a
|
serine/arginine-rich splicing factor 1a |
| chr19_-_18689571 | 0.43 |
ENSDART00000016135
|
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr25_-_35820568 | 0.42 |
ENSDART00000152766
|
CR354435.4
|
Histone H2B 1/2 |
| chr20_+_52778914 | 0.41 |
ENSDART00000138641
|
si:dkey-235d18.5
|
si:dkey-235d18.5 |
| chr19_-_18315998 | 0.41 |
ENSDART00000151133
|
top2b
|
topoisomerase (DNA) II beta |
| chr19_+_13492428 | 0.41 |
ENSDART00000165033
|
srsf4
|
serine/arginine-rich splicing factor 4 |
| chr19_-_3208863 | 0.40 |
|
|
|
| chr23_+_44782492 | 0.40 |
ENSDART00000125198
|
pfn1
|
profilin 1 |
| chr17_-_20216303 | 0.39 |
ENSDART00000029380
|
bnip4
|
BCL2/adenovirus E1B interacting protein 4 |
| KN149932v1_+_27584 | 0.39 |
|
|
|
| chr7_-_6323808 | 0.39 |
ENSDART00000173158
|
zgc:112234
|
zgc:112234 |
| chr11_-_18016658 | 0.39 |
|
|
|
| chr11_+_14263376 | 0.39 |
ENSDART00000132997
|
ptbp1b
|
polypyrimidine tract binding protein 1b |
| chr3_+_15355472 | 0.38 |
ENSDART00000141714
|
nupr1
|
nuclear protein 1 |
| chr11_-_2227358 | 0.38 |
ENSDART00000018131
|
hnrnpa1a
|
heterogeneous nuclear ribonucleoprotein A1a |
| chr14_-_50145578 | 0.38 |
ENSDART00000042421
|
ppp2ca
|
protein phosphatase 2, catalytic subunit, alpha isozyme |
| chr20_-_25731057 | 0.38 |
|
|
|
| chr18_-_38235232 | 0.37 |
ENSDART00000114224
|
si:dkey-10o6.2
|
si:dkey-10o6.2 |
| chr16_-_36061564 | 0.36 |
ENSDART00000175847
|
CABZ01065723.1
|
ENSDARG00000105999 |
| chr7_-_71290605 | 0.36 |
ENSDART00000111797
|
acer2
|
alkaline ceramidase 2 |
| chr2_+_39653315 | 0.36 |
ENSDART00000077108
|
zgc:136870
|
zgc:136870 |
| chr14_-_38487071 | 0.36 |
ENSDART00000160000
|
spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr6_-_9471808 | 0.36 |
ENSDART00000039280
|
fzd7b
|
frizzled class receptor 7b |
| chr7_-_57030751 | 0.35 |
|
|
|
| chr20_+_25669615 | 0.35 |
ENSDART00000063107
|
cyp2p7
|
cytochrome P450, family 2, subfamily P, polypeptide 7 |
| chr5_+_69401770 | 0.35 |
|
|
|
| chr4_+_33077499 | 0.34 |
|
|
|
| chr19_+_42863138 | 0.34 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr22_+_2681009 | 0.34 |
|
|
|
| chr13_-_7897936 | 0.34 |
ENSDART00000139728
|
si:ch211-250c4.4
|
si:ch211-250c4.4 |
| chr9_-_19951894 | 0.33 |
ENSDART00000146841
|
si:ch211-141e20.2
|
si:ch211-141e20.2 |
| chr6_-_120518 | 0.33 |
ENSDART00000148911
|
keap1b
|
kelch-like ECH-associated protein 1b |
| chr17_-_14697094 | 0.33 |
ENSDART00000154281
|
ptp4a2a
|
protein tyrosine phosphatase type IVA, member 2a |
| chr12_-_20493683 | 0.33 |
|
|
|
| chr25_+_35784213 | 0.33 |
ENSDART00000152761
|
CR354435.7
|
ENSDARG00000094744 |
| chr19_+_15617624 | 0.32 |
ENSDART00000003164
|
ppp1r8a
|
protein phosphatase 1, regulatory subunit 8a |
| chr11_-_17620732 | 0.32 |
ENSDART00000154627
|
eogt
|
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr18_+_38793474 | 0.32 |
ENSDART00000059208
|
fam214a
|
family with sequence similarity 214, member A |
| chr7_+_71262935 | 0.32 |
ENSDART00000100396
|
ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr3_+_62051743 | 0.32 |
ENSDART00000168063
|
zgc:173575
|
zgc:173575 |
| chr2_+_2930605 | 0.32 |
ENSDART00000124882
|
thoc1
|
THO complex 1 |
| chr19_+_43448344 | 0.31 |
ENSDART00000015632
|
nkain1
|
sodium/potassium transporting ATPase interacting 1 |
| chr5_+_69096325 | 0.31 |
ENSDART00000014649
|
ugt2a5
|
UDP glucuronosyltransferase 2 family, polypeptide A5 |
| chr3_+_62081343 | 0.31 |
ENSDART00000058719
|
znf1124
|
zinc finger protein 1124 |
| chr9_-_9264634 | 0.31 |
ENSDART00000021191
|
u2af1
|
U2 small nuclear RNA auxiliary factor 1 |
| chr19_-_29205158 | 0.30 |
ENSDART00000026992
|
sox4a
|
SRY (sex determining region Y)-box 4a |
| chr21_-_767364 | 0.30 |
ENSDART00000130648
|
scarb2c
|
scavenger receptor class B, member 2c |
| chr19_+_19330545 | 0.30 |
ENSDART00000157523
|
slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr9_-_23346038 | 0.30 |
ENSDART00000135461
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr16_+_51319421 | 0.30 |
|
|
|
| chr20_+_49975268 | 0.30 |
ENSDART00000008808
|
hmbox1b
|
homeobox containing 1 b |
| chr14_+_10350292 | 0.29 |
ENSDART00000127594
|
atrx
|
alpha thalassemia/mental retardation syndrome X-linked homolog (human) |
| chr7_+_20008083 | 0.29 |
ENSDART00000012450
|
dvl2
|
dishevelled segment polarity protein 2 |
| chr8_+_8934762 | 0.28 |
ENSDART00000133137
|
bcap31
|
B-cell receptor-associated protein 31 |
| chr18_+_40535215 | 0.28 |
ENSDART00000132833
|
vwa7
|
von Willebrand factor A domain containing 7 |
| chr7_+_40879017 | 0.28 |
|
|
|
| chr15_+_43192811 | 0.28 |
|
|
|
| chr5_+_64411584 | 0.27 |
ENSDART00000158484
|
BX001023.2
|
ENSDARG00000101163 |
| chr19_+_20199890 | 0.27 |
ENSDART00000161019
|
hoxa4a
|
homeobox A4a |
| chr24_+_14095643 | 0.27 |
ENSDART00000124740
|
ncoa2
|
nuclear receptor coactivator 2 |
| chr9_-_39190534 | 0.27 |
|
|
|
| chr14_+_6122644 | 0.26 |
ENSDART00000149783
ENSDART00000148461 |
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr11_+_37453275 | 0.26 |
ENSDART00000050296
|
snrpe
|
small nuclear ribonucleoprotein polypeptide E |
| chr21_-_28883441 | 0.26 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr18_-_36291301 | 0.25 |
|
|
|
| chr4_+_29775735 | 0.25 |
ENSDART00000166510
|
BX530097.1
|
ENSDARG00000104762 |
| chr21_+_37405904 | 0.24 |
ENSDART00000148956
|
znf346
|
zinc finger protein 346 |
| chr18_+_41537683 | 0.24 |
ENSDART00000146972
|
selt1b
|
selenoprotein T, 1b |
| chr16_-_31664756 | 0.24 |
ENSDART00000176939
|
phf20l1
|
PHD finger protein 20-like 1 |
| chr24_+_16402613 | 0.24 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr16_-_33151504 | 0.24 |
ENSDART00000145055
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr21_-_25704793 | 0.23 |
ENSDART00000101211
|
cldnh
|
claudin h |
| chr23_+_2391317 | 0.23 |
|
|
|
| chr10_-_43815475 | 0.23 |
ENSDART00000027392
|
mef2ca
|
myocyte enhancer factor 2ca |
| chr13_+_25298383 | 0.23 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0071887 | positive regulation of adaptive immune response(GO:0002821) positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains(GO:0002824) negative regulation of phospholipase activity(GO:0010519) positive regulation of vesicle fusion(GO:0031340) type 2 immune response(GO:0042092) regulation of fatty acid biosynthetic process(GO:0042304) T-helper 2 cell differentiation(GO:0045064) leukocyte apoptotic process(GO:0071887) regulation of leukocyte apoptotic process(GO:2000106) |
| 0.5 | 1.9 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.5 | 5.1 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.4 | 1.3 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.3 | 2.4 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.3 | 0.9 | GO:0071947 | protein K29-linked deubiquitination(GO:0035523) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.2 | 1.7 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.2 | 0.8 | GO:2000009 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.2 | 0.7 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.2 | 0.5 | GO:0016121 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.1 | 0.7 | GO:1903963 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.1 | 0.8 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.1 | 2.4 | GO:0007097 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.1 | 1.2 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.1 | 0.8 | GO:0090179 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 4.3 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.1 | 1.4 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.1 | 0.7 | GO:0003315 | heart rudiment formation(GO:0003315) |
| 0.1 | 0.5 | GO:0038026 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 0.4 | GO:0071459 | protein localization to kinetochore(GO:0034501) protein localization to chromosome, centromeric region(GO:0071459) |
| 0.1 | 0.3 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.3 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.1 | 0.3 | GO:0010922 | positive regulation of phosphatase activity(GO:0010922) positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 1.0 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 0.2 | GO:0001502 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.1 | 0.2 | GO:0044806 | multicellular organism aging(GO:0010259) G-quadruplex DNA unwinding(GO:0044806) |
| 0.1 | 1.3 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.1 | 0.5 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.1 | 0.8 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 0.2 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.1 | 0.2 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.1 | 0.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.9 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 1.2 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.1 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.4 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 2.4 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.6 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.0 | 0.5 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.1 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.5 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.6 | GO:0009409 | response to cold(GO:0009409) positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.2 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.4 | GO:1902110 | positive regulation of mitochondrial membrane permeability(GO:0035794) mitochondrial outer membrane permeabilization(GO:0097345) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.0 | 1.2 | GO:0006305 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.9 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 1.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.1 | GO:0031294 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.3 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.2 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.1 | GO:0072526 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.3 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.3 | GO:0071577 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.2 | GO:0050829 | defense response to Gram-negative bacterium(GO:0050829) |
| 0.0 | 0.7 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.3 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.1 | GO:0045887 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) positive regulation of neuromuscular junction development(GO:1904398) |
| 0.0 | 0.4 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.1 | GO:0051029 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.0 | 0.2 | GO:0009743 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.0 | 0.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.3 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 1.4 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 1.4 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 0.3 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.8 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.1 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 0.6 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.7 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.4 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.3 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.1 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.2 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.3 | 2.7 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.2 | 2.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 0.9 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 1.1 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 1.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.4 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.1 | 1.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.3 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 1.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 6.5 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.5 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.5 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 0.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.3 | 2.7 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.3 | 4.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 1.6 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.2 | 0.5 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.1 | 0.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 1.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.5 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 0.3 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.4 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 0.7 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 0.8 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.1 | 0.3 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 1.0 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 1.6 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 1.2 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.9 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 3.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 1.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.3 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 2.3 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.1 | 0.8 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.2 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 2.4 | GO:0045499 | semaphorin receptor binding(GO:0030215) chemorepellent activity(GO:0045499) |
| 0.0 | 0.3 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.6 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 1.0 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.1 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 2.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.7 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 1.9 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 0.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 0.7 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.9 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.8 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.7 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.4 | 2.4 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 1.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.1 | 1.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 0.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.8 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.8 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.7 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.3 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |