Project
DANIO-CODE
Navigation
Downloads
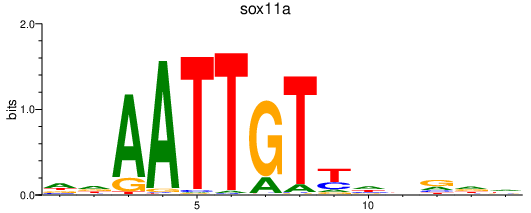
Results for sox11a
Z-value: 0.32
Transcription factors associated with sox11a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox11a
|
ENSDARG00000077811 | SRY-box transcription factor 11a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox11a | dr10_dc_chr17_-_35934175_35934284 | 0.04 | 8.9e-01 | Click! |
Activity profile of sox11a motif
Sorted Z-values of sox11a motif
Network of associatons between targets according to the STRING database.
First level regulatory network of sox11a
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_60973097 | 0.36 |
ENSDART00000055062
|
pvalb1
|
parvalbumin 1 |
| chr21_-_41445163 | 0.30 |
ENSDART00000170457
|
hmp19
|
HMP19 protein |
| chr21_+_26352675 | 0.29 |
ENSDART00000077197
|
tmsb
|
thymosin, beta |
| chr25_+_34551602 | 0.29 |
ENSDART00000121825
|
hist1h4l
|
histone 1, H4, like |
| chr11_+_40652239 | 0.27 |
ENSDART00000169817
ENSDART00000162157 ENSDART00000172008 |
pax7a
|
paired box 7a |
| chr7_+_58427742 | 0.25 |
ENSDART00000073640
|
plag1
|
pleiomorphic adenoma gene 1 |
| chr15_+_19536380 | 0.24 |
ENSDART00000154779
|
zgc:77784
|
zgc:77784 |
| chr24_+_21476448 | 0.22 |
ENSDART00000002595
|
rpl21
|
ribosomal protein L21 |
| chr5_+_68943914 | 0.21 |
ENSDART00000159594
|
mthfd2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr3_+_15245529 | 0.21 |
|
|
|
| chr22_-_36552513 | 0.20 |
ENSDART00000129318
|
CABZ01045212.1
|
ENSDARG00000087525 |
| chr22_+_17581859 | 0.20 |
ENSDART00000035670
|
polr2eb
|
polymerase (RNA) II (DNA directed) polypeptide E, b |
| KN150698v1_+_1663 | 0.20 |
ENSDART00000163899
|
ENSDARG00000100052
|
ENSDARG00000100052 |
| chr6_+_14822990 | 0.19 |
ENSDART00000149949
|
pou3f3b
|
POU class 3 homeobox 3b |
| chr11_-_29910947 | 0.19 |
ENSDART00000156121
|
scml2
|
sex comb on midleg-like 2 (Drosophila) |
| chr1_-_13547500 | 0.19 |
ENSDART00000044896
|
camk2d2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II delta 2 |
| chr6_+_45493396 | 0.19 |
ENSDART00000159863
|
cntn4
|
contactin 4 |
| KN149959v1_-_18526 | 0.18 |
ENSDART00000178715
|
CABZ01078607.1
|
ENSDARG00000109077 |
| chr25_+_34553079 | 0.18 |
ENSDART00000156838
|
zgc:112234
|
zgc:112234 |
| chr19_-_47897712 | 0.17 |
|
|
|
| chr6_+_14823185 | 0.17 |
ENSDART00000149202
|
pou3f3b
|
POU class 3 homeobox 3b |
| chr19_+_27133834 | 0.17 |
ENSDART00000134455
|
zgc:100906
|
zgc:100906 |
| chr14_+_7596207 | 0.16 |
ENSDART00000113299
|
zgc:110843
|
zgc:110843 |
| chr1_-_33981055 | 0.16 |
ENSDART00000102111
|
tpt1
|
tumor protein, translationally-controlled 1 |
| chr9_+_33167554 | 0.16 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr16_-_24682800 | 0.16 |
ENSDART00000156407
ENSDART00000154072 |
si:dkey-56f14.4
|
si:dkey-56f14.4 |
| chr12_-_30897542 | 0.15 |
ENSDART00000145967
|
tcf7l2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr18_-_43890514 | 0.15 |
ENSDART00000087382
|
ddx6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr9_+_51693805 | 0.15 |
ENSDART00000132896
|
fap
|
fibroblast activation protein, alpha |
| chr22_+_16282153 | 0.15 |
ENSDART00000162685
ENSDART00000105678 |
lrrc39
|
leucine rich repeat containing 39 |
| chr17_-_43041127 | 0.15 |
ENSDART00000132754
|
npc2
|
Niemann-Pick disease, type C2 |
| chr19_+_24334831 | 0.15 |
ENSDART00000090200
|
snapin
|
SNAP-associated protein |
| chr2_+_24521514 | 0.14 |
ENSDART00000099546
|
map4l
|
microtubule associated protein 4 like |
| chr20_-_5184885 | 0.14 |
ENSDART00000114851
|
ccdc85cb
|
coiled-coil domain containing 85C, b |
| chr17_+_45430353 | 0.14 |
ENSDART00000162937
|
ezra
|
ezrin a |
| chr15_-_1858350 | 0.14 |
ENSDART00000082026
|
mmp28
|
matrix metallopeptidase 28 |
| chr16_+_46239355 | 0.14 |
ENSDART00000168145
|
bola1
|
bolA family member 1 |
| chr18_-_25190648 | 0.14 |
ENSDART00000175178
|
slco3a1
|
solute carrier organic anion transporter family, member 3A1 |
| chr14_+_4044357 | 0.13 |
ENSDART00000160431
|
dhrs13l1
|
dehydrogenase/reductase (SDR family) member 13 like 1 |
| chr4_+_14107347 | 0.13 |
ENSDART00000150748
|
CU469420.1
|
ENSDARG00000086259 |
| chr17_+_24421150 | 0.13 |
ENSDART00000168926
|
mdh1ab
|
malate dehydrogenase 1Ab, NAD (soluble) |
| chr15_-_15421065 | 0.13 |
ENSDART00000106120
|
ywhag2
|
3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide 2 |
| chr24_-_37407313 | 0.12 |
|
|
|
| chr10_+_21693418 | 0.12 |
ENSDART00000160754
|
pcdh1g1
|
protocadherin 1 gamma 1 |
| chr5_-_14330177 | 0.12 |
|
|
|
| chr9_-_18561215 | 0.12 |
ENSDART00000084668
|
enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr19_-_9603597 | 0.12 |
ENSDART00000045245
|
ing4
|
inhibitor of growth family, member 4 |
| chr23_-_22724076 | 0.12 |
ENSDART00000132733
|
ca6
|
carbonic anhydrase VI |
| chr18_+_9524331 | 0.11 |
ENSDART00000053125
|
sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr9_+_6684835 | 0.11 |
|
|
|
| chr13_-_42630425 | 0.11 |
ENSDART00000161950
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr4_-_71770315 | 0.11 |
ENSDART00000171434
|
zgc:162958
|
zgc:162958 |
| chr2_+_38390887 | 0.11 |
ENSDART00000113111
|
psmb5
|
proteasome subunit beta 5 |
| chr12_-_36294319 | 0.11 |
ENSDART00000157746
ENSDART00000166925 |
pde6g
|
phosphodiesterase 6G, cGMP-specific, rod, gamma |
| chr20_-_16271651 | 0.11 |
ENSDART00000012476
|
coa7
|
cytochrome c oxidase assembly factor 7 |
| chr2_+_28688137 | 0.11 |
ENSDART00000169614
|
nadsyn1
|
NAD synthetase 1 |
| chr4_+_14827397 | 0.11 |
ENSDART00000158686
|
si:dkey-14k9.2
|
si:dkey-14k9.2 |
| chr20_-_48255596 | 0.11 |
ENSDART00000031167
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr20_+_51034244 | 0.11 |
|
|
|
| chr10_-_27047692 | 0.11 |
ENSDART00000139942
ENSDART00000146983 |
uqcc3
|
ubiquinol-cytochrome c reductase complex assembly factor 3 |
| chr23_+_31888903 | 0.11 |
ENSDART00000075730
|
myb
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr13_-_25067585 | 0.10 |
ENSDART00000159585
|
adka
|
adenosine kinase a |
| chr9_-_31467299 | 0.10 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr10_+_22754558 | 0.10 |
ENSDART00000136123
|
kdm6bb
|
lysine (K)-specific demethylase 6B, b |
| chr14_-_29487265 | 0.10 |
|
|
|
| chr20_+_36098607 | 0.10 |
ENSDART00000021456
|
opn5
|
opsin 5 |
| chr17_-_39237265 | 0.09 |
ENSDART00000050534
|
crim1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
| chr9_+_2001849 | 0.09 |
ENSDART00000082329
|
evx2
|
even-skipped homeobox 2 |
| chr5_+_25604463 | 0.09 |
ENSDART00000137178
|
marveld2b
|
MARVEL domain containing 2b |
| chr21_+_41684338 | 0.09 |
ENSDART00000169511
|
ppp2r2bb
|
protein phosphatase 2, regulatory subunit B, beta b |
| chr2_+_4534596 | 0.09 |
ENSDART00000157744
ENSDART00000163111 ENSDART00000163986 |
wacb
|
WW domain containing adaptor with coiled-coil b |
| chr6_-_31268267 | 0.09 |
|
|
|
| chr17_-_43040958 | 0.09 |
ENSDART00000132754
|
npc2
|
Niemann-Pick disease, type C2 |
| chr1_-_18809429 | 0.09 |
ENSDART00000124260
|
kitb
|
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog b |
| chr6_-_50705420 | 0.09 |
ENSDART00000074100
|
osgn1
|
oxidative stress induced growth inhibitor 1 |
| KN150127v1_-_16267 | 0.08 |
|
|
|
| chr12_-_18286666 | 0.08 |
ENSDART00000078767
|
tom1l2
|
target of myb1 like 2 membrane trafficking protein |
| chr5_-_31328823 | 0.08 |
ENSDART00000142919
|
ssh1b
|
slingshot protein phosphatase 1b |
| chr1_-_54087595 | 0.08 |
ENSDART00000150362
|
si:ch211-197k17.3
|
si:ch211-197k17.3 |
| chr5_+_24039974 | 0.08 |
ENSDART00000029699
|
atp6v0a2b
|
ATPase, H+ transporting, lysosomal V0 subunit a2b |
| chr3_+_29849717 | 0.07 |
ENSDART00000151509
|
knca7
|
potassium voltage-gated channel, shaker-related subfamily, member 7 |
| chr7_-_39874079 | 0.07 |
ENSDART00000173634
|
wdr60
|
WD repeat domain 60 |
| chr25_+_34553195 | 0.07 |
ENSDART00000156838
|
zgc:112234
|
zgc:112234 |
| chr2_+_52639037 | 0.07 |
ENSDART00000006216
|
plpp2a
|
phospholipid phosphatase 2a |
| chr18_+_16203004 | 0.07 |
ENSDART00000135016
|
lrriq1
|
leucine-rich repeats and IQ motif containing 1 |
| chr1_-_28849479 | 0.07 |
ENSDART00000109224
|
ENSDARG00000074117
|
ENSDARG00000074117 |
| chr18_+_7632469 | 0.07 |
ENSDART00000136313
|
si:dkeyp-1h4.6
|
si:dkeyp-1h4.6 |
| chr10_+_24690680 | 0.07 |
ENSDART00000147974
|
vps36
|
vacuolar protein sorting 36 homolog (S. cerevisiae) |
| chr18_+_23891152 | 0.06 |
ENSDART00000146490
|
BX569787.1
|
ENSDARG00000092402 |
| chr2_-_40170303 | 0.06 |
ENSDART00000165602
|
epha4a
|
eph receptor A4a |
| chr19_-_34508557 | 0.06 |
ENSDART00000160495
|
si:dkey-184p18.2
|
si:dkey-184p18.2 |
| chr5_-_68943600 | 0.06 |
ENSDART00000159851
|
mob1a
|
MOB kinase activator 1A |
| chr13_-_42630594 | 0.06 |
ENSDART00000111255
|
capn2a
|
calpain 2, (m/II) large subunit a |
| chr15_+_17164535 | 0.06 |
ENSDART00000155350
|
relb
|
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr19_-_5453469 | 0.06 |
ENSDART00000105004
|
krt17
|
keratin 17 |
| chr16_+_2661916 | 0.06 |
ENSDART00000149675
ENSDART00000109980 |
lars2
|
leucyl-tRNA synthetase 2, mitochondrial |
| chr20_-_31164373 | 0.06 |
ENSDART00000014163
|
fndc1
|
fibronectin type III domain containing 1 |
| chr3_+_21925465 | 0.06 |
|
|
|
| chr16_-_42015262 | 0.06 |
ENSDART00000136742
|
BX649388.1
|
ENSDARG00000091933 |
| chr13_+_33497110 | 0.06 |
ENSDART00000026464
|
cfl1l
|
cofilin 1 (non-muscle), like |
| chr14_-_2268924 | 0.06 |
ENSDART00000160205
|
pcdh2ab12
|
protocadherin 2 alpha b 12 |
| chr15_+_35395257 | 0.06 |
ENSDART00000066737
|
zgc:158328
|
zgc:158328 |
| chr8_-_39944817 | 0.06 |
ENSDART00000083066
|
asphd2
|
aspartate beta-hydroxylase domain containing 2 |
| chr5_-_67233396 | 0.06 |
ENSDART00000051833
ENSDART00000124890 |
gsx1
|
GS homeobox 1 |
| chr13_+_13562712 | 0.06 |
ENSDART00000110509
ENSDART00000169265 |
si:ch211-194c3.5
|
si:ch211-194c3.5 |
| chr9_+_3414293 | 0.06 |
|
|
|
| chr19_-_35828807 | 0.06 |
|
|
|
| chr10_+_2559774 | 0.06 |
ENSDART00000126937
|
wu:fb59d01
|
wu:fb59d01 |
| chr9_+_24198112 | 0.06 |
ENSDART00000148226
|
mlphb
|
melanophilin b |
| chr8_-_12171624 | 0.06 |
ENSDART00000132824
|
dab2ipa
|
DAB2 interacting protein a |
| chr16_-_36085222 | 0.05 |
ENSDART00000159134
|
OSCP1
|
organic solute carrier partner 1 |
| chr17_+_33344090 | 0.05 |
ENSDART00000123114
|
dnajc27
|
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr18_-_47186896 | 0.05 |
|
|
|
| chr19_-_9603672 | 0.05 |
ENSDART00000045245
|
ing4
|
inhibitor of growth family, member 4 |
| chr2_-_44519346 | 0.05 |
|
|
|
| chr16_+_23172295 | 0.05 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
| chr15_+_29842298 | 0.05 |
|
|
|
| chr5_+_26195283 | 0.05 |
ENSDART00000053001
|
tcn2
|
transcobalamin II |
| chr22_-_17571930 | 0.05 |
|
|
|
| chr6_+_14823900 | 0.05 |
ENSDART00000178555
|
CU929521.3
|
ENSDARG00000108533 |
| chr20_+_54463948 | 0.05 |
ENSDART00000158810
ENSDART00000161631 ENSDART00000172631 ENSDART00000168924 |
fkbp3
|
FK506 binding protein 3 |
| chr17_+_45430228 | 0.05 |
ENSDART00000162937
|
ezra
|
ezrin a |
| chr22_+_2253153 | 0.05 |
ENSDART00000133751
|
AL929017.2
|
ENSDARG00000095449 |
| chr20_+_33972327 | 0.05 |
ENSDART00000170930
|
rxrgb
|
retinoid X receptor, gamma b |
| chr25_-_14327948 | 0.05 |
ENSDART00000073609
|
prmt7
|
protein arginine methyltransferase 7 |
| chr16_-_17164459 | 0.05 |
ENSDART00000178443
|
tnfrsf1a
|
tumor necrosis factor receptor superfamily, member 1a |
| chr8_-_16689642 | 0.05 |
ENSDART00000049676
|
depdc1a
|
DEP domain containing 1a |
| chr21_-_38106145 | 0.05 |
ENSDART00000151226
|
klf5l
|
Kruppel-like factor 5 like |
| chr5_-_34701639 | 0.04 |
ENSDART00000085142
|
map1b
|
microtubule-associated protein 1B |
| chr25_-_3521317 | 0.04 |
ENSDART00000171863
ENSDART00000166363 |
si:ch211-272n13.3
|
si:ch211-272n13.3 |
| chr17_+_33388313 | 0.04 |
ENSDART00000077553
|
xdh
|
xanthine dehydrogenase |
| chr3_+_18657831 | 0.04 |
ENSDART00000055757
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr10_-_27047617 | 0.04 |
ENSDART00000139942
|
uqcc3
|
ubiquinol-cytochrome c reductase complex assembly factor 3 |
| chr17_-_52735615 | 0.04 |
|
|
|
| chr9_-_21687972 | 0.04 |
ENSDART00000131634
|
aff3
|
AF4/FMR2 family, member 3 |
| chr5_+_61281695 | 0.04 |
ENSDART00000134344
|
si:dkeyp-117b8.4
|
si:dkeyp-117b8.4 |
| chr12_-_37560192 | 0.04 |
ENSDART00000140353
|
sdk2b
|
sidekick cell adhesion molecule 2b |
| KN149959v1_+_21015 | 0.03 |
ENSDART00000161495
|
CABZ01078608.1
|
ENSDARG00000105028 |
| chr23_-_25760788 | 0.03 |
ENSDART00000088208
|
lrp1ab
|
low density lipoprotein receptor-related protein 1Ab |
| chr6_+_50452529 | 0.03 |
ENSDART00000155051
|
mych
|
myelocytomatosis oncogene homolog |
| chr22_-_23518719 | 0.03 |
ENSDART00000161253
|
crb1
|
crumbs family member 1, photoreceptor morphogenesis associated |
| chr18_+_9413668 | 0.03 |
ENSDART00000061886
|
sema3ab
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Ab |
| chr24_-_32259029 | 0.03 |
ENSDART00000048633
|
trdmt1
|
tRNA aspartic acid methyltransferase 1 |
| chr14_-_25637853 | 0.03 |
ENSDART00000139855
|
g3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr18_-_8422039 | 0.03 |
ENSDART00000141581
|
sephs1
|
selenophosphate synthetase 1 |
| chr5_-_24039681 | 0.03 |
ENSDART00000046384
|
trmt2a
|
tRNA methyltransferase 2 homolog A |
| chr1_+_4704661 | 0.03 |
ENSDART00000040204
|
tuba8l2
|
tubulin, alpha 8 like 2 |
| chr5_+_9579430 | 0.03 |
ENSDART00000109236
|
ENSDARG00000075416
|
ENSDARG00000075416 |
| chr2_-_2456819 | 0.02 |
ENSDART00000013794
ENSDART00000082155 ENSDART00000169871 ENSDART00000167202 |
dab1b
|
Dab, reelin signal transducer, homolog 1b (Drosophila) |
| chr11_-_865070 | 0.02 |
ENSDART00000172904
|
atg7
|
ATG7 autophagy related 7 homolog (S. cerevisiae) |
| chr6_+_4068830 | 0.02 |
ENSDART00000130642
|
ENSDARG00000088225
|
ENSDARG00000088225 |
| chr2_-_30071872 | 0.02 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr6_-_37766424 | 0.02 |
ENSDART00000149722
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr11_-_1382526 | 0.02 |
ENSDART00000121537
|
si:ch211-266k22.6
|
si:ch211-266k22.6 |
| chr19_-_34508527 | 0.02 |
ENSDART00000160495
|
si:dkey-184p18.2
|
si:dkey-184p18.2 |
| chr17_+_45430125 | 0.02 |
ENSDART00000124911
|
ezra
|
ezrin a |
| chr11_+_25358389 | 0.02 |
ENSDART00000140856
|
ccdc120
|
coiled-coil domain containing 120 |
| chr8_-_1833095 | 0.02 |
ENSDART00000114476
ENSDART00000091235 ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr8_+_1052572 | 0.02 |
ENSDART00000081445
|
ubox5
|
U-box domain containing 5 |
| chr1_-_25751157 | 0.02 |
|
|
|
| chr20_+_19894077 | 0.02 |
ENSDART00000136317
|
chrna2b
|
cholinergic receptor, nicotinic, alpha 2b (neuronal) |
| chr9_+_19031775 | 0.02 |
ENSDART00000055866
|
chmp2ba
|
charged multivesicular body protein 2Ba |
| chr20_+_9141086 | 0.02 |
ENSDART00000064144
ENSDART00000144396 |
bpnt1
|
bisphosphate nucleotidase 1 |
| chr19_+_44288538 | 0.01 |
ENSDART00000139684
|
lypla2
|
lysophospholipase II |
| chr4_+_27139902 | 0.01 |
|
|
|
| chr2_+_26585236 | 0.01 |
ENSDART00000129895
|
palm1b
|
paralemmin 1b |
| chr8_+_5222145 | 0.01 |
ENSDART00000035676
|
bnip3la
|
BCL2/adenovirus E1B interacting protein 3-like a |
| chr24_+_24999138 | 0.01 |
ENSDART00000152095
ENSDART00000154988 ENSDART00000156098 |
CR383672.2
|
ENSDARG00000096452 |
| chr21_-_30217568 | 0.01 |
ENSDART00000101054
|
dnajc18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr4_+_12980657 | 0.01 |
|
|
|
| chr5_-_60747054 | 0.01 |
ENSDART00000082952
|
rcc1l
|
RCC1 like |
| chr5_-_68943821 | 0.01 |
ENSDART00000158956
|
mob1a
|
MOB kinase activator 1A |
| chr11_+_30266605 | 0.01 |
ENSDART00000008594
|
tmem178
|
transmembrane protein 178 |
| chr10_+_42530040 | 0.01 |
ENSDART00000025691
|
dbnla
|
drebrin-like a |
| chr12_-_24729980 | 0.01 |
|
|
|
| chr2_-_23003738 | 0.01 |
ENSDART00000115025
|
thap4
|
THAP domain containing 4 |
| chr8_+_52544578 | 0.01 |
ENSDART00000127729
|
stambpb
|
STAM binding protein b |
| chr21_+_26576104 | 0.01 |
ENSDART00000142667
|
esrra
|
estrogen-related receptor alpha |
| chr11_-_865163 | 0.01 |
ENSDART00000172904
|
atg7
|
ATG7 autophagy related 7 homolog (S. cerevisiae) |
| chr11_-_1382493 | 0.01 |
ENSDART00000121537
|
si:ch211-266k22.6
|
si:ch211-266k22.6 |
| chr10_+_41746636 | 0.01 |
ENSDART00000127073
|
lrrc75bb
|
leucine rich repeat containing 75Bb |
| chr2_-_30071815 | 0.01 |
ENSDART00000056747
|
shhb
|
sonic hedgehog b |
| chr24_-_41357096 | 0.01 |
ENSDART00000150207
|
acvr2b
|
activin A receptor, type IIB |
| chr4_+_28385257 | 0.01 |
ENSDART00000076037
|
alg10
|
asparagine-linked glycosylation 10 |
| chr5_-_41396258 | 0.00 |
ENSDART00000143573
|
ncor1
|
nuclear receptor corepressor 1 |
| chr10_+_24690784 | 0.00 |
ENSDART00000147974
|
vps36
|
vacuolar protein sorting 36 homolog (S. cerevisiae) |
| chr11_-_40632136 | 0.00 |
ENSDART00000163609
|
mrps16
|
mitochondrial ribosomal protein S16 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0019730 | antimicrobial humoral response(GO:0019730) antibacterial humoral response(GO:0019731) |
| 0.1 | 0.3 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.0 | 0.1 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0007624 | ultradian rhythm(GO:0007624) |
| 0.0 | 0.2 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.0 | 0.1 | GO:0030852 | regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.0 | 0.2 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.2 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0021521 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.0 | 0.1 | GO:0038093 | B cell differentiation(GO:0030183) Fc receptor signaling pathway(GO:0038093) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.1 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0031083 | BLOC-1 complex(GO:0031083) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.2 | GO:0004488 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.0 | 0.1 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.1 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0016918 | retinal binding(GO:0016918) |
| 0.0 | 0.0 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.0 | GO:0030151 | oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0038191 | neuropilin binding(GO:0038191) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.1 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |