Project
DANIO-CODE
Navigation
Downloads
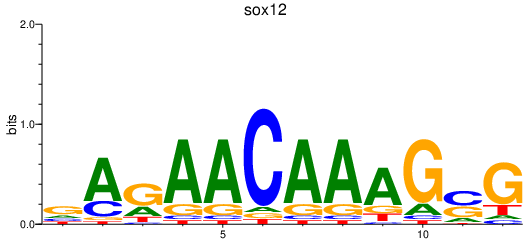
Results for sox12
Z-value: 0.33
Transcription factors associated with sox12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox12
|
ENSDARG00000025847 | SRY-box transcription factor 12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox12 | dr10_dc_chr11_-_23954234_23954320 | -0.47 | 6.6e-02 | Click! |
Activity profile of sox12 motif
Sorted Z-values of sox12 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of sox12
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_32716794 | 0.34 |
ENSDART00000141013
|
atg101
|
autophagy related 101 |
| chr20_-_14218080 | 0.30 |
ENSDART00000104032
|
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr17_-_23689233 | 0.29 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr20_+_1364879 | 0.27 |
ENSDART00000145981
ENSDART00000152709 |
mmachc
|
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria |
| chr17_+_7377230 | 0.27 |
ENSDART00000157123
|
shprh
|
SNF2 histone linker PHD RING helicase |
| chr7_-_24249672 | 0.26 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr8_+_54034258 | 0.26 |
ENSDART00000170712
ENSDART00000164153 |
BRPF1
|
bromodomain and PHD finger containing 1 |
| chr7_-_28293919 | 0.25 |
ENSDART00000028887
|
tmem9b
|
TMEM9 domain family, member B |
| chr2_-_31849916 | 0.24 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr12_-_22279843 | 0.23 |
ENSDART00000153194
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr6_+_29763568 | 0.22 |
ENSDART00000151784
|
ptmaa
|
prothymosin, alpha a |
| chr22_-_746251 | 0.22 |
ENSDART00000061775
|
rbbp5
|
retinoblastoma binding protein 5 |
| chr5_-_23727236 | 0.22 |
ENSDART00000143492
|
senp3a
|
SUMO1/sentrin/SMT3 specific peptidase 3a |
| chr16_-_16329761 | 0.22 |
ENSDART00000111912
|
rbm12b
|
RNA binding motif protein 12B |
| chr2_-_56933166 | 0.22 |
ENSDART00000167441
|
ARMC6
|
armadillo repeat containing 6 |
| chr14_+_45685886 | 0.22 |
|
|
|
| chr24_-_41403824 | 0.21 |
ENSDART00000063504
ENSDART00000169374 |
xylb
|
xylulokinase homolog (H. influenzae) |
| chr3_-_15960288 | 0.20 |
ENSDART00000051807
|
lasp1
|
LIM and SH3 protein 1 |
| chr16_-_10006640 | 0.20 |
ENSDART00000104058
|
ncalda
|
neurocalcin delta a |
| chr24_-_2453890 | 0.20 |
ENSDART00000093331
|
rreb1a
|
ras responsive element binding protein 1a |
| chr7_-_24724911 | 0.20 |
ENSDART00000173791
|
rcor2
|
REST corepressor 2 |
| chr18_+_8954323 | 0.20 |
ENSDART00000134827
|
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr6_-_12787256 | 0.19 |
ENSDART00000157139
|
tmbim1a
|
transmembrane BAX inhibitor motif containing 1a |
| chr22_+_21593142 | 0.19 |
ENSDART00000133939
|
tle2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr3_-_15960491 | 0.19 |
ENSDART00000064838
|
lasp1
|
LIM and SH3 protein 1 |
| chr8_-_19283046 | 0.18 |
ENSDART00000164780
|
rgl1
|
ral guanine nucleotide dissociation stimulator-like 1 |
| chr24_+_37810878 | 0.18 |
ENSDART00000066556
|
zmp:0000000624
|
zmp:0000000624 |
| chr17_-_25284710 | 0.18 |
ENSDART00000167992
|
ppie
|
peptidylprolyl isomerase E (cyclophilin E) |
| chr18_+_6339577 | 0.18 |
ENSDART00000136333
|
wash1
|
WAS protein family homolog 1 |
| chr14_-_21320696 | 0.17 |
ENSDART00000043162
|
reep2
|
receptor accessory protein 2 |
| chr5_-_23727210 | 0.17 |
ENSDART00000143492
|
senp3a
|
SUMO1/sentrin/SMT3 specific peptidase 3a |
| chr3_+_10182687 | 0.17 |
ENSDART00000156700
|
cbx2
|
chromobox homolog 2 (Drosophila Pc class) |
| chr10_-_32716718 | 0.16 |
ENSDART00000063544
|
atg101
|
autophagy related 101 |
| chr17_+_51851738 | 0.16 |
ENSDART00000053422
|
ttll5
|
tubulin tyrosine ligase-like family, member 5 |
| chr4_-_3014778 | 0.16 |
ENSDART00000128934
ENSDART00000019518 |
aebp2
|
AE binding protein 2 |
| chr4_-_17028492 | 0.16 |
|
|
|
| chr20_+_14218237 | 0.15 |
ENSDART00000044937
|
kcns3b
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3b |
| chr16_+_12922320 | 0.15 |
ENSDART00000008535
|
u2af2a
|
U2 small nuclear RNA auxiliary factor 2a |
| chr18_+_8954512 | 0.14 |
ENSDART00000134827
|
tmem243a
|
transmembrane protein 243, mitochondrial a |
| chr13_+_35639424 | 0.14 |
ENSDART00000100156
|
agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr3_+_10182994 | 0.14 |
ENSDART00000156700
|
cbx2
|
chromobox homolog 2 (Drosophila Pc class) |
| chr22_+_21593704 | 0.14 |
|
|
|
| chr15_+_27000467 | 0.14 |
ENSDART00000023842
|
ppm1da
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Da |
| chr16_+_12922781 | 0.13 |
ENSDART00000008535
|
u2af2a
|
U2 small nuclear RNA auxiliary factor 2a |
| chr3_+_40142679 | 0.13 |
ENSDART00000009411
|
bud31
|
BUD31 homolog (S. cerevisiae) |
| chr16_-_26633959 | 0.13 |
ENSDART00000139663
|
CR391910.2
|
ENSDARG00000093592 |
| chr22_-_746441 | 0.13 |
ENSDART00000061775
|
rbbp5
|
retinoblastoma binding protein 5 |
| chr3_-_35734943 | 0.13 |
ENSDART00000135957
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr17_-_25284780 | 0.12 |
ENSDART00000167992
|
ppie
|
peptidylprolyl isomerase E (cyclophilin E) |
| chr6_-_12787356 | 0.12 |
ENSDART00000157139
|
tmbim1a
|
transmembrane BAX inhibitor motif containing 1a |
| chr17_+_30826890 | 0.12 |
ENSDART00000149600
|
tpp1
|
tripeptidyl peptidase I |
| chr17_-_23689380 | 0.12 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr12_-_22279890 | 0.12 |
ENSDART00000153194
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr17_-_23689317 | 0.12 |
ENSDART00000124661
|
papss2a
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2a |
| chr17_+_15528691 | 0.12 |
|
|
|
| chr3_-_35735315 | 0.12 |
ENSDART00000102952
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr14_-_49973159 | 0.12 |
ENSDART00000124192
|
si:dkeyp-121d2.7
|
si:dkeyp-121d2.7 |
| chr2_-_31850133 | 0.12 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr7_+_28844711 | 0.11 |
ENSDART00000052345
|
tradd
|
tnfrsf1a-associated via death domain |
| chr5_-_37784663 | 0.11 |
ENSDART00000051233
|
mink1
|
misshapen-like kinase 1 |
| chr17_-_7377094 | 0.11 |
|
|
|
| chr1_+_40784540 | 0.11 |
ENSDART00000142508
|
si:ch211-89o9.6
|
si:ch211-89o9.6 |
| chr18_+_14368973 | 0.11 |
|
|
|
| chr23_+_30803887 | 0.11 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr14_-_6637670 | 0.10 |
ENSDART00000126279
|
clk4a
|
CDC-like kinase 4a |
| chr2_-_56933194 | 0.10 |
ENSDART00000178179
|
ARMC6
|
armadillo repeat containing 6 |
| chr5_-_27549500 | 0.10 |
ENSDART00000051515
|
zgc:110329
|
zgc:110329 |
| chr2_-_24413154 | 0.10 |
ENSDART00000145526
|
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr7_-_24249630 | 0.10 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr22_-_746334 | 0.10 |
ENSDART00000061775
|
rbbp5
|
retinoblastoma binding protein 5 |
| chr10_+_5267937 | 0.09 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr8_+_54034431 | 0.09 |
ENSDART00000170712
ENSDART00000164153 |
BRPF1
|
bromodomain and PHD finger containing 1 |
| chr6_+_46404956 | 0.09 |
ENSDART00000131203
ENSDART00000103472 ENSDART00000132845 ENSDART00000138567 ENSDART00000168440 |
pbrm1l
|
polybromo 1, like |
| chr15_-_28654137 | 0.09 |
ENSDART00000156049
|
ssh2a
|
slingshot protein phosphatase 2a |
| chr5_+_25620803 | 0.09 |
ENSDART00000007587
|
gtf2h2
|
general transcription factor IIH, polypeptide 2 |
| chr22_+_39012456 | 0.08 |
ENSDART00000176985
|
CABZ01044615.1
|
ENSDARG00000108810 |
| chr9_-_34145854 | 0.08 |
|
|
|
| chr21_-_26369503 | 0.08 |
ENSDART00000137312
|
eif4ebp3
|
eukaryotic translation initiation factor 4E binding protein 3 |
| chr7_-_24724452 | 0.08 |
ENSDART00000173791
|
rcor2
|
REST corepressor 2 |
| chr6_-_12786937 | 0.08 |
ENSDART00000157139
|
tmbim1a
|
transmembrane BAX inhibitor motif containing 1a |
| chr12_-_22279713 | 0.07 |
ENSDART00000153194
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr10_+_5268009 | 0.07 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr24_+_26223767 | 0.07 |
|
|
|
| chr2_-_56933086 | 0.07 |
ENSDART00000167441
|
ARMC6
|
armadillo repeat containing 6 |
| chr20_+_31372117 | 0.07 |
|
|
|
| chr6_-_55287399 | 0.07 |
|
|
|
| chr23_+_30803518 | 0.07 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr1_-_25838798 | 0.07 |
|
|
|
| chr5_+_44538762 | 0.07 |
ENSDART00000172702
ENSDART00000136002 |
smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr7_-_28293954 | 0.06 |
ENSDART00000028887
|
tmem9b
|
TMEM9 domain family, member B |
| chr6_+_27676886 | 0.06 |
ENSDART00000159624
ENSDART00000049177 |
rab6ba
|
RAB6B, member RAS oncogene family a |
| chr9_+_25109716 | 0.06 |
ENSDART00000037025
|
slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr1_-_33694144 | 0.06 |
|
|
|
| chr4_+_28374965 | 0.06 |
ENSDART00000150424
|
si:ch73-263o4.4
|
si:ch73-263o4.4 |
| chr20_-_14218236 | 0.06 |
ENSDART00000168434
|
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr15_+_27000248 | 0.06 |
ENSDART00000023842
|
ppm1da
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Da |
| chr9_-_46609555 | 0.05 |
|
|
|
| chr8_-_9531866 | 0.05 |
ENSDART00000044000
|
plxna3
|
plexin A3 |
| chr23_-_36349563 | 0.05 |
|
|
|
| chr6_-_11594818 | 0.05 |
ENSDART00000151602
|
AL929243.1
|
ENSDARG00000096290 |
| chr13_+_35639263 | 0.05 |
ENSDART00000100156
|
agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr13_+_4040744 | 0.05 |
|
|
|
| chr17_+_23102283 | 0.04 |
ENSDART00000138069
|
rasgrp3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr17_+_15528825 | 0.04 |
|
|
|
| chr11_+_23657658 | 0.04 |
ENSDART00000160394
|
CR354444.2
|
ENSDARG00000103549 |
| chr5_-_68712571 | 0.03 |
ENSDART00000109487
|
CABZ01032476.1
|
ENSDARG00000074645 |
| chr7_-_26653629 | 0.03 |
ENSDART00000124363
|
alx4a
|
ALX homeobox 4a |
| chr7_+_38395803 | 0.03 |
ENSDART00000013394
|
mtch2
|
mitochondrial carrier homolog 2 |
| chr6_+_46404650 | 0.03 |
ENSDART00000131203
ENSDART00000103472 ENSDART00000132845 ENSDART00000138567 ENSDART00000168440 |
pbrm1l
|
polybromo 1, like |
| chr24_-_37793048 | 0.03 |
ENSDART00000078828
ENSDART00000131342 |
anks3
|
ankyrin repeat and sterile alpha motif domain containing 3 |
| chr22_+_21593358 | 0.03 |
ENSDART00000133939
|
tle2
|
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
| chr23_-_36350288 | 0.03 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr23_-_26799063 | 0.02 |
ENSDART00000157844
|
hspg2
|
heparan sulfate proteoglycan 2 |
| chr3_-_35735085 | 0.02 |
ENSDART00000135957
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr17_+_7377152 | 0.02 |
ENSDART00000157123
|
shprh
|
SNF2 histone linker PHD RING helicase |
| chr17_+_23102259 | 0.02 |
ENSDART00000178403
|
rasgrp3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr19_-_3208863 | 0.02 |
|
|
|
| chr17_-_37040694 | 0.01 |
ENSDART00000126823
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr5_-_68712851 | 0.01 |
ENSDART00000109487
|
CABZ01032476.1
|
ENSDARG00000074645 |
| chr13_+_9227395 | 0.01 |
ENSDART00000047740
|
wdr32
|
WD repeat domain 32 |
| chr18_+_44635321 | 0.01 |
|
|
|
| chr9_-_33116897 | 0.01 |
|
|
|
| chr7_-_26653672 | 0.01 |
ENSDART00000124363
|
alx4a
|
ALX homeobox 4a |
| chr15_+_27000663 | 0.01 |
ENSDART00000023842
|
ppm1da
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Da |
| chr18_-_14368801 | 0.01 |
ENSDART00000164100
|
hsbp1b
|
heat shock factor binding protein 1b |
| chr4_-_13568618 | 0.00 |
ENSDART00000169582
|
mdm1
|
Mdm1 nuclear protein homolog (mouse) |
| chr1_+_45817971 | 0.00 |
|
|
|
| chr15_-_1519571 | 0.00 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0050427 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.1 | 0.2 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.4 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.3 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 0.2 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.2 | GO:0046660 | female sex differentiation(GO:0046660) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.3 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.0 | 0.2 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 0.1 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.2 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.0 | 0.5 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.3 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.4 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.2 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.3 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.2 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.4 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.2 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.3 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 0.2 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |