Project
DANIO-CODE
Navigation
Downloads
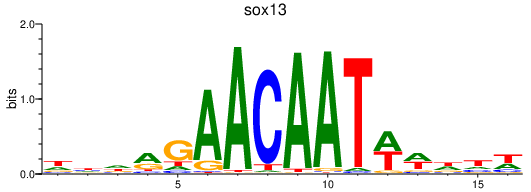
Results for sox13
Z-value: 0.82
Transcription factors associated with sox13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox13
|
ENSDARG00000030297 | SRY-box transcription factor 13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox13 | dr10_dc_chr11_+_37501309_37501381 | -0.59 | 1.5e-02 | Click! |
Activity profile of sox13 motif
Sorted Z-values of sox13 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of sox13
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_4835526 | 1.66 |
|
|
|
| chr2_+_38019437 | 1.29 |
ENSDART00000139564
|
dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr9_+_2523927 | 1.28 |
ENSDART00000166326
|
si:ch73-167c12.2
|
si:ch73-167c12.2 |
| chr3_+_42383724 | 1.24 |
|
|
|
| chr12_-_48759104 | 1.22 |
ENSDART00000130190
ENSDART00000105309 |
uros
|
uroporphyrinogen III synthase |
| chr4_-_65423861 | 1.21 |
ENSDART00000175471
|
BX511215.1
|
ENSDARG00000098062 |
| chr4_-_35856278 | 1.16 |
ENSDART00000162568
|
znf1125
|
zinc finger protein 1125 |
| chr12_-_22279843 | 1.16 |
ENSDART00000153194
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr5_+_27793171 | 1.16 |
|
|
|
| chr13_+_22973764 | 1.15 |
ENSDART00000110266
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr13_+_11303809 | 1.12 |
ENSDART00000169895
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr7_-_55346967 | 1.12 |
ENSDART00000135304
|
pabpn1l
|
poly(A) binding protein, nuclear 1-like (cytoplasmic) |
| chr25_+_5845303 | 1.08 |
ENSDART00000163948
|
ENSDARG00000053246
|
ENSDARG00000053246 |
| chr14_+_15845853 | 1.01 |
ENSDART00000160973
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr16_-_55211053 | 1.00 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr6_-_46733412 | 0.98 |
ENSDART00000103455
|
tarbp2
|
TAR (HIV) RNA binding protein 2 |
| chr20_-_34126039 | 0.97 |
ENSDART00000033817
|
scyl3
|
SCY1-like, kinase-like 3 |
| chr16_-_43107682 | 0.96 |
ENSDART00000142003
|
nudt17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr5_+_24039736 | 0.92 |
ENSDART00000175389
|
atp6v0a2b
|
ATPase, H+ transporting, lysosomal V0 subunit a2b |
| chr10_+_24530670 | 0.89 |
|
|
|
| chr24_+_29902165 | 0.88 |
ENSDART00000168422
ENSDART00000100092 ENSDART00000113304 |
frrs1b
|
ferric-chelate reductase 1b |
| chr7_-_26226451 | 0.87 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr2_+_34984631 | 0.86 |
ENSDART00000141796
|
astn1
|
astrotactin 1 |
| chr14_+_15845943 | 0.85 |
ENSDART00000159352
|
polr1a
|
polymerase (RNA) I polypeptide A |
| chr12_-_22279890 | 0.84 |
ENSDART00000153194
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr17_-_7575628 | 0.84 |
ENSDART00000064657
|
stx11a
|
syntaxin 11a |
| chr19_-_35286549 | 0.83 |
ENSDART00000016057
|
ctnnal1
|
catenin (cadherin-associated protein), alpha-like 1 |
| chr12_+_29121368 | 0.83 |
ENSDART00000006505
|
mxtx2
|
mix-type homeobox gene 2 |
| chr15_-_34107138 | 0.83 |
ENSDART00000163841
|
lsr
|
lipolysis stimulated lipoprotein receptor |
| chr16_+_7463067 | 0.83 |
ENSDART00000149086
|
atg5
|
ATG5 autophagy related 5 homolog (S. cerevisiae) |
| chr2_-_57339717 | 0.82 |
ENSDART00000150034
|
pias4b
|
protein inhibitor of activated STAT, 4b |
| chr11_-_6979695 | 0.82 |
ENSDART00000173242
ENSDART00000172896 ENSDART00000102493 |
zgc:173548
|
zgc:173548 |
| chr5_-_8712114 | 0.82 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr10_-_21587697 | 0.82 |
ENSDART00000029122
|
zgc:165539
|
zgc:165539 |
| chr8_-_39788989 | 0.81 |
ENSDART00000067843
|
zgc:162025
|
zgc:162025 |
| chr15_+_40228037 | 0.79 |
ENSDART00000111018
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr11_+_25302044 | 0.79 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr1_-_29958364 | 0.79 |
ENSDART00000085454
|
dis3
|
DIS3 exosome endoribonuclease and 3'-5' exoribonuclease |
| chr5_-_8206288 | 0.78 |
ENSDART00000062957
|
paip1
|
poly(A) binding protein interacting protein 1 |
| chr15_-_20189604 | 0.78 |
ENSDART00000152355
|
med13b
|
mediator complex subunit 13b |
| chr14_-_23813378 | 0.77 |
ENSDART00000133522
|
cpeb4a
|
cytoplasmic polyadenylation element binding protein 4a |
| chr19_+_7489293 | 0.76 |
ENSDART00000081746
|
apoa1bp
|
apolipoprotein A-I binding protein |
| chr11_+_1558144 | 0.73 |
ENSDART00000154583
|
si:dkey-40c23.2
|
si:dkey-40c23.2 |
| chr19_-_24333844 | 0.72 |
ENSDART00000138279
|
chtopa
|
chromatin target of PRMT1a |
| chr1_-_343261 | 0.72 |
ENSDART00000010092
|
gas6
|
growth arrest-specific 6 |
| chr5_-_24039681 | 0.72 |
ENSDART00000046384
|
trmt2a
|
tRNA methyltransferase 2 homolog A |
| chr13_-_49934440 | 0.72 |
ENSDART00000034541
|
gpatch11
|
G patch domain containing 11 |
| chr8_-_1833095 | 0.71 |
ENSDART00000114476
ENSDART00000091235 ENSDART00000140077 |
pi4kab
|
phosphatidylinositol 4-kinase, catalytic, alpha b |
| chr20_+_35535503 | 0.71 |
ENSDART00000153249
|
tdrd6
|
tudor domain containing 6 |
| chr14_-_47210912 | 0.71 |
ENSDART00000031640
ENSDART00000132966 |
zgc:113425
|
zgc:113425 |
| chr15_-_34733101 | 0.70 |
ENSDART00000003674
|
ankmy2a
|
ankyrin repeat and MYND domain containing 2a |
| chr3_+_32440178 | 0.70 |
ENSDART00000151025
|
si:ch73-248e21.1
|
si:ch73-248e21.1 |
| chr2_+_37224986 | 0.70 |
ENSDART00000138952
|
apoda.2
|
apolipoprotein Da, duplicate 2 |
| chr14_-_7437682 | 0.70 |
ENSDART00000172215
|
si:ch211-39i2.2
|
si:ch211-39i2.2 |
| KN150339v1_-_39357 | 0.69 |
ENSDART00000169638
|
wu:fa19b12
|
wu:fa19b12 |
| chr1_+_25750825 | 0.68 |
ENSDART00000046376
|
g3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr18_+_8959686 | 0.68 |
ENSDART00000145226
|
si:ch211-233h19.2
|
si:ch211-233h19.2 |
| chr5_-_8712068 | 0.68 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr3_+_32278978 | 0.67 |
ENSDART00000061426
|
rras
|
related RAS viral (r-ras) oncogene homolog |
| chr17_-_6225136 | 0.67 |
ENSDART00000137389
ENSDART00000115389 |
txlnbb
|
taxilin beta b |
| chr2_+_30756519 | 0.67 |
ENSDART00000101861
|
tcea1
|
transcription elongation factor A (SII), 1 |
| chr5_+_12090143 | 0.66 |
ENSDART00000133167
|
si:dkey-98f17.5
|
si:dkey-98f17.5 |
| chr21_+_26954898 | 0.66 |
ENSDART00000114469
|
fkbp2
|
FK506 binding protein 2 |
| chr14_+_35084220 | 0.66 |
ENSDART00000171809
|
sytl4
|
synaptotagmin-like 4 |
| chr2_+_38019262 | 0.65 |
ENSDART00000178610
|
dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr15_+_22458649 | 0.65 |
ENSDART00000109931
|
oafa
|
OAF homolog a (Drosophila) |
| chr2_+_38019379 | 0.65 |
ENSDART00000139564
|
dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr22_-_25015224 | 0.65 |
ENSDART00000015512
|
polr3f
|
polymerase (RNA) III (DNA directed) polypeptide F |
| chr7_-_24724911 | 0.64 |
ENSDART00000173791
|
rcor2
|
REST corepressor 2 |
| chr18_-_3430582 | 0.64 |
ENSDART00000169049
|
capn5a
|
calpain 5a |
| chr3_+_32279283 | 0.62 |
ENSDART00000061426
|
rras
|
related RAS viral (r-ras) oncogene homolog |
| chr3_+_56882443 | 0.62 |
|
|
|
| chr8_-_51380691 | 0.61 |
ENSDART00000060628
|
chmp7
|
charged multivesicular body protein 7 |
| chr18_-_7488960 | 0.61 |
ENSDART00000052803
|
si:dkey-30c15.10
|
si:dkey-30c15.10 |
| chr8_+_11287550 | 0.61 |
ENSDART00000115057
|
tjp2b
|
tight junction protein 2b (zona occludens 2) |
| chr14_+_30434751 | 0.61 |
ENSDART00000139975
|
atl3
|
atlastin 3 |
| chr16_-_25318413 | 0.60 |
ENSDART00000058943
|
ENSDARG00000040280
|
ENSDARG00000040280 |
| chr10_-_21404605 | 0.60 |
ENSDART00000125167
|
avd
|
avidin |
| chr16_+_45370954 | 0.59 |
ENSDART00000147343
|
dnase2
|
deoxyribonuclease II, lysosomal |
| chr2_+_24982203 | 0.59 |
ENSDART00000088346
|
mast3a
|
microtubule associated serine/threonine kinase 3a |
| chr23_-_17055508 | 0.58 |
ENSDART00000125472
|
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr9_-_34450885 | 0.58 |
ENSDART00000012385
|
me3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr11_+_25302180 | 0.57 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr6_+_44199738 | 0.57 |
ENSDART00000075486
|
ppp4r2b
|
protein phosphatase 4, regulatory subunit 2b |
| chr10_+_33758581 | 0.57 |
ENSDART00000141650
|
b3glctb
|
beta 3-glucosyltransferase b |
| chr22_+_2483845 | 0.56 |
ENSDART00000170192
|
CU570894.1
|
ENSDARG00000103886 |
| chr22_-_6532901 | 0.56 |
ENSDART00000106100
|
zgc:171490
|
zgc:171490 |
| chr6_-_52677363 | 0.56 |
ENSDART00000083830
|
sdc4
|
syndecan 4 |
| chr5_+_9579430 | 0.56 |
ENSDART00000109236
|
ENSDARG00000075416
|
ENSDARG00000075416 |
| chr12_-_13511174 | 0.55 |
ENSDART00000133895
|
ghdc
|
GH3 domain containing |
| chr5_-_8206388 | 0.55 |
ENSDART00000062957
|
paip1
|
poly(A) binding protein interacting protein 1 |
| chr5_+_51979524 | 0.55 |
ENSDART00000168317
|
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr24_+_24025923 | 0.55 |
ENSDART00000127842
|
si:dkey-226l10.6
|
si:dkey-226l10.6 |
| chr13_-_33186642 | 0.54 |
ENSDART00000110295
|
tmem234
|
transmembrane protein 234 |
| chr4_+_48372465 | 0.54 |
|
|
|
| chr7_-_71343686 | 0.54 |
ENSDART00000128678
|
mettl4
|
methyltransferase like 4 |
| chr11_+_8142982 | 0.54 |
ENSDART00000166379
|
samd13
|
sterile alpha motif domain containing 13 |
| chr21_-_20908880 | 0.54 |
ENSDART00000079701
|
rnf180
|
ring finger protein 180 |
| chr18_+_24490621 | 0.54 |
ENSDART00000143108
|
CR847971.1
|
ENSDARG00000091941 |
| chr10_-_17593407 | 0.53 |
ENSDART00000145077
|
ska1
|
spindle and kinetochore associated complex subunit 1 |
| chr19_-_17306473 | 0.53 |
ENSDART00000007906
|
stmn1a
|
stathmin 1a |
| chr11_-_25223594 | 0.53 |
ENSDART00000014945
|
hcfc1a
|
host cell factor C1a |
| chr9_+_32838316 | 0.53 |
ENSDART00000172033
|
rabl3
|
RAB, member of RAS oncogene family-like 3 |
| chr11_-_14044356 | 0.52 |
ENSDART00000085158
|
tmem259
|
transmembrane protein 259 |
| chr17_+_16184621 | 0.52 |
ENSDART00000156832
|
kif13ba
|
kinesin family member 13Ba |
| chr18_-_24490329 | 0.51 |
ENSDART00000147961
|
CR847971.2
|
ENSDARG00000094853 |
| chr24_+_36429525 | 0.51 |
ENSDART00000062722
|
pus3
|
pseudouridylate synthase 3 |
| chr3_+_39413882 | 0.51 |
ENSDART00000156848
|
BX537342.1
|
ENSDARG00000097765 |
| chr24_-_41275160 | 0.51 |
|
|
|
| chr15_-_14162320 | 0.51 |
ENSDART00000156947
|
si:ch211-116o3.5
|
si:ch211-116o3.5 |
| chr14_+_35065189 | 0.50 |
ENSDART00000171565
|
zbtb3
|
zinc finger and BTB domain containing 3 |
| KN150030v1_-_22613 | 0.50 |
ENSDART00000175410
|
CABZ01079802.1
|
ENSDARG00000106760 |
| chr21_-_25358498 | 0.50 |
ENSDART00000167189
|
ppme1
|
protein phosphatase methylesterase 1 |
| chr13_+_35346924 | 0.50 |
ENSDART00000011583
|
mkks
|
McKusick-Kaufman syndrome |
| chr14_-_30604897 | 0.50 |
ENSDART00000161540
|
si:zfos-80g12.1
|
si:zfos-80g12.1 |
| chr14_-_15777250 | 0.49 |
ENSDART00000165656
|
mxd3
|
MAX dimerization protein 3 |
| chr25_-_21684852 | 0.49 |
ENSDART00000089616
|
bmt2
|
base methyltransferase of 25S rRNA 2 homolog |
| chr5_-_23079460 | 0.49 |
ENSDART00000027217
|
cwc15
|
CWC15 spliceosome-associated protein homolog (S. cerevisiae) |
| chr24_+_29902377 | 0.49 |
ENSDART00000168422
ENSDART00000100092 ENSDART00000113304 |
frrs1b
|
ferric-chelate reductase 1b |
| chr2_-_31850133 | 0.49 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr23_-_17055330 | 0.49 |
ENSDART00000125472
|
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr4_-_20119812 | 0.48 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr5_-_8206177 | 0.48 |
ENSDART00000062957
|
paip1
|
poly(A) binding protein interacting protein 1 |
| chr2_-_32530224 | 0.48 |
ENSDART00000170674
|
abcf2a
|
ATP-binding cassette, sub-family F (GCN20), member 2a |
| chr23_-_17055376 | 0.48 |
ENSDART00000125472
|
dnmt3bb.3
|
DNA (cytosine-5-)-methyltransferase beta, duplicate b.3 |
| chr21_+_11742219 | 0.48 |
ENSDART00000081661
|
nudt2
|
nudix (nucleoside diphosphate linked moiety X)-type motif 2 |
| chr2_-_31849916 | 0.47 |
ENSDART00000109460
|
nol7
|
nucleolar protein 7 |
| chr9_-_40863087 | 0.47 |
ENSDART00000135134
|
bard1
|
BRCA1 associated RING domain 1 |
| chr2_+_27674700 | 0.47 |
ENSDART00000087643
|
tesk2
|
testis-specific kinase 2 |
| chr3_-_36298676 | 0.47 |
ENSDART00000157950
|
rogdi
|
rogdi homolog (Drosophila) |
| chr17_+_11921923 | 0.47 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr4_+_17653548 | 0.47 |
ENSDART00000026509
|
cwf19l1
|
CWF19-like 1, cell cycle control |
| chr14_-_22198294 | 0.47 |
ENSDART00000137167
|
ENSDARG00000056830
|
ENSDARG00000056830 |
| chr2_+_38019291 | 0.46 |
ENSDART00000139564
|
dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr16_+_17857553 | 0.46 |
ENSDART00000148878
|
them4
|
thioesterase superfamily member 4 |
| chr16_-_44179964 | 0.46 |
ENSDART00000058685
|
zfpm2a
|
zinc finger protein, FOG family member 2a |
| chr1_-_21021029 | 0.45 |
ENSDART00000129066
|
zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr3_+_56882467 | 0.45 |
|
|
|
| chr5_-_23291839 | 0.45 |
ENSDART00000099083
ENSDART00000099084 |
gbgt1l4
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 4 |
| chr4_-_43539382 | 0.45 |
|
|
|
| chr5_-_28914505 | 0.45 |
ENSDART00000098364
|
ehmt1a
|
euchromatic histone-lysine N-methyltransferase 1a |
| chr18_-_12358605 | 0.45 |
ENSDART00000114024
|
fam107b
|
family with sequence similarity 107, member B |
| chr19_-_41820114 | 0.44 |
ENSDART00000038038
|
shfm1
|
split hand/foot malformation (ectrodactyly) type 1 |
| chr1_+_25751216 | 0.44 |
ENSDART00000046376
|
g3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr13_-_44645490 | 0.44 |
ENSDART00000099990
|
btbd9
|
BTB (POZ) domain containing 9 |
| chr13_-_25590425 | 0.44 |
ENSDART00000142404
|
si:dkey-192p21.6
|
si:dkey-192p21.6 |
| chr14_-_40941231 | 0.43 |
ENSDART00000138917
|
arl13a
|
ADP-ribosylation factor-like 13A |
| chr14_-_20940726 | 0.43 |
ENSDART00000129743
|
si:ch211-175m2.5
|
si:ch211-175m2.5 |
| chr12_-_20536385 | 0.43 |
ENSDART00000080888
|
snf8
|
SNF8, ESCRT-II complex subunit, homolog (S. cerevisiae) |
| chr17_-_11312603 | 0.43 |
ENSDART00000091159
|
adpgk2
|
ADP-dependent glucokinase 2 |
| chr5_+_24039974 | 0.43 |
ENSDART00000029699
|
atp6v0a2b
|
ATPase, H+ transporting, lysosomal V0 subunit a2b |
| chr7_-_26226924 | 0.43 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr9_+_38672040 | 0.43 |
ENSDART00000157556
|
lss
|
lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) |
| chr2_-_22836607 | 0.43 |
ENSDART00000168022
|
gtf2b
|
general transcription factor IIB |
| chr1_+_25750998 | 0.43 |
ENSDART00000046376
|
g3bp2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr3_+_52290252 | 0.42 |
ENSDART00000018908
|
slc27a1a
|
solute carrier family 27 (fatty acid transporter), member 1a |
| chr2_-_44330306 | 0.42 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr15_+_1018191 | 0.42 |
|
|
|
| chr5_+_53633496 | 0.42 |
ENSDART00000166050
|
rangrf
|
RAN guanine nucleotide release factor |
| chr3_-_26675055 | 0.41 |
ENSDART00000143710
|
pigq
|
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr24_-_24884787 | 0.41 |
ENSDART00000168254
|
nup58
|
nucleoporin 58 |
| chr5_+_21673232 | 0.41 |
ENSDART00000131223
|
fdx1b
|
ferredoxin 1b |
| chr13_+_45387478 | 0.41 |
ENSDART00000019113
|
tmem57b
|
transmembrane protein 57b |
| chr5_-_8206218 | 0.41 |
ENSDART00000062957
|
paip1
|
poly(A) binding protein interacting protein 1 |
| chr10_-_28494204 | 0.41 |
ENSDART00000131220
|
btg3
|
B-cell translocation gene 3 |
| chr2_+_24982268 | 0.41 |
ENSDART00000088346
|
mast3a
|
microtubule associated serine/threonine kinase 3a |
| chr7_+_32626520 | 0.41 |
ENSDART00000140800
ENSDART00000075263 ENSDART00000137956 |
tssc4
|
tumor suppressing subtransferable candidate 4 |
| chr14_+_7596207 | 0.40 |
ENSDART00000113299
|
zgc:110843
|
zgc:110843 |
| chr3_+_32279635 | 0.40 |
ENSDART00000141793
|
rras
|
related RAS viral (r-ras) oncogene homolog |
| chr1_-_30301568 | 0.40 |
ENSDART00000132466
|
si:ch211-269i23.2
|
si:ch211-269i23.2 |
| chr5_-_25133229 | 0.40 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr9_-_12472519 | 0.39 |
ENSDART00000102434
|
ehhadh
|
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| chr24_+_36451347 | 0.39 |
ENSDART00000142264
|
grnb
|
granulin b |
| chr19_-_17306333 | 0.39 |
ENSDART00000132662
|
stmn1a
|
stathmin 1a |
| chr13_-_31267459 | 0.39 |
ENSDART00000008287
|
pgam1a
|
phosphoglycerate mutase 1a |
| chr19_-_18199573 | 0.39 |
ENSDART00000166313
|
thrb
|
thyroid hormone receptor beta |
| chr23_-_1548272 | 0.39 |
ENSDART00000160184
ENSDART00000136436 |
epm2a
|
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr11_+_25302072 | 0.39 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr16_-_34523796 | 0.39 |
ENSDART00000136546
|
serinc2l
|
serine incorporator 2, like |
| chr14_+_47131478 | 0.39 |
ENSDART00000017785
|
naa15a
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit a |
| chr8_+_32380573 | 0.39 |
ENSDART00000146901
|
mthfd2l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr11_-_23458624 | 0.39 |
ENSDART00000124810
|
pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 beta |
| chr15_+_40227972 | 0.38 |
ENSDART00000111018
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr5_+_56539999 | 0.38 |
ENSDART00000167660
|
pja2
|
praja ring finger 2 |
| chr3_+_48527525 | 0.38 |
ENSDART00000149755
|
tyk2
|
tyrosine kinase 2 |
| chr6_-_46733353 | 0.38 |
ENSDART00000103455
|
tarbp2
|
TAR (HIV) RNA binding protein 2 |
| chr6_-_6138267 | 0.38 |
ENSDART00000092172
|
mtif2
|
mitochondrial translational initiation factor 2 |
| chr18_-_49291686 | 0.38 |
ENSDART00000174038
|
si:zfos-464b6.2
|
si:zfos-464b6.2 |
| chr23_+_23305376 | 0.37 |
ENSDART00000126479
|
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr20_+_44600015 | 0.37 |
ENSDART00000023763
|
wdcp
|
WD repeat and coiled coil containing |
| chr14_+_29945396 | 0.37 |
ENSDART00000173090
|
mtus1a
|
microtubule associated tumor suppressor 1a |
| chr23_-_18781059 | 0.37 |
ENSDART00000144668
|
ENSDARG00000019503
|
ENSDARG00000019503 |
| chr14_-_32937341 | 0.36 |
ENSDART00000048130
|
stard14
|
START domain containing 14 |
| chr14_+_11872906 | 0.36 |
ENSDART00000054626
|
hdac3
|
histone deacetylase 3 |
| chr23_+_2105366 | 0.36 |
|
|
|
| chr8_-_22720007 | 0.36 |
|
|
|
| chr12_-_14964222 | 0.36 |
ENSDART00000108852
|
pkmyt1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr5_-_22364806 | 0.36 |
ENSDART00000057541
|
apool
|
apolipoprotein O-like |
| chr10_-_14971565 | 0.35 |
ENSDART00000147653
|
smad2
|
SMAD family member 2 |
| chr13_-_17333287 | 0.35 |
ENSDART00000145499
|
c13h10orf11
|
c13h10orf11 homolog (H. sapiens) |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:1903798 | regulation of symbiosis, encompassing mutualism through parasitism(GO:0043903) regulation of production of small RNA involved in gene silencing by RNA(GO:0070920) small RNA loading onto RISC(GO:0070922) regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903798) |
| 0.3 | 1.0 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.3 | 1.2 | GO:0048103 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 0.3 | 1.0 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.2 | 0.7 | GO:0001659 | medium-chain fatty acid transport(GO:0001579) temperature homeostasis(GO:0001659) |
| 0.2 | 0.8 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.2 | 0.8 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.2 | 0.6 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.2 | 1.8 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.2 | 1.8 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.4 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.1 | 0.9 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.1 | 0.8 | GO:0030262 | apoptotic DNA fragmentation(GO:0006309) cellular component disassembly involved in execution phase of apoptosis(GO:0006921) apoptotic nuclear changes(GO:0030262) |
| 0.1 | 0.5 | GO:1903059 | regulation of lipoprotein metabolic process(GO:0050746) regulation of protein lipidation(GO:1903059) |
| 0.1 | 0.7 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.1 | 0.4 | GO:0046552 | thyroid hormone mediated signaling pathway(GO:0002154) retinal cone cell fate determination(GO:0042671) eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate determination(GO:0043703) retinal cone cell fate commitment(GO:0046551) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 1.7 | GO:0035090 | maintenance of cell polarity(GO:0030011) maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.5 | GO:1990481 | tRNA pseudouridine synthesis(GO:0031119) mRNA pseudouridine synthesis(GO:1990481) |
| 0.1 | 1.4 | GO:0051452 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 0.5 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.8 | GO:0097065 | anterior head development(GO:0097065) |
| 0.1 | 0.4 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 0.3 | GO:0045813 | positive regulation of Wnt signaling pathway, calcium modulating pathway(GO:0045813) |
| 0.1 | 0.3 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.1 | 0.4 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.1 | 0.8 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.1 | 0.4 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.1 | 0.4 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 0.6 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.3 | GO:0070849 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.1 | 0.3 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.1 | 0.6 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 1.0 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.1 | 0.6 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.5 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.1 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) actin filament debranching(GO:0071846) |
| 0.1 | 0.3 | GO:0040014 | regulation of multicellular organism growth(GO:0040014) |
| 0.1 | 0.5 | GO:0009712 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.1 | 0.2 | GO:0031548 | neural plate formation(GO:0021990) brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) positive regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031550) anterior neural plate formation(GO:0090017) |
| 0.1 | 0.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.1 | 0.6 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.1 | 0.5 | GO:1904292 | regulation of ERAD pathway(GO:1904292) |
| 0.1 | 0.2 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.1 | 0.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.1 | 0.7 | GO:0030719 | P granule organization(GO:0030719) |
| 0.1 | 2.4 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.1 | 0.4 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.8 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.1 | 0.2 | GO:1904396 | regulation of neuromuscular junction development(GO:1904396) |
| 0.1 | 0.5 | GO:0001840 | neural plate development(GO:0001840) |
| 0.1 | 0.4 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.2 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.1 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.2 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.2 | GO:0018410 | C-terminal protein amino acid modification(GO:0018410) |
| 0.0 | 0.1 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.4 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.0 | 0.4 | GO:0031113 | regulation of microtubule polymerization(GO:0031113) |
| 0.0 | 0.5 | GO:0006167 | AMP biosynthetic process(GO:0006167) ATP biosynthetic process(GO:0006754) |
| 0.0 | 1.0 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.2 | GO:0042987 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.0 | 0.5 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.6 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.3 | GO:0031106 | septin ring organization(GO:0031106) septin cytoskeleton organization(GO:0032185) |
| 0.0 | 0.1 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.4 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.4 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.4 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.2 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.0 | 0.7 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.2 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.3 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 1.6 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0009186 | purine deoxyribonucleotide metabolic process(GO:0009151) purine deoxyribonucleotide catabolic process(GO:0009155) deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.2 | GO:0099517 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.1 | GO:0033572 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.0 | 0.2 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.2 | GO:0000022 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.7 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 0.4 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) |
| 0.0 | 0.3 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.2 | GO:0033505 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.3 | GO:0021684 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 2.0 | GO:0021782 | glial cell development(GO:0021782) |
| 0.0 | 2.0 | GO:0007492 | endoderm development(GO:0007492) |
| 0.0 | 0.2 | GO:0019856 | pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 0.4 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.0 | 0.4 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.3 | GO:0071156 | regulation of cell cycle arrest(GO:0071156) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.4 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.2 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.5 | GO:0039021 | pronephric glomerulus development(GO:0039021) |
| 0.0 | 0.7 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.1 | GO:0045950 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.3 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.3 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.2 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 1.1 | GO:0006096 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 | 0.5 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.3 | GO:0060541 | respiratory system development(GO:0060541) |
| 0.0 | 1.5 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.1 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) dendritic cell differentiation(GO:0097028) |
| 0.0 | 0.9 | GO:0008589 | regulation of smoothened signaling pathway(GO:0008589) |
| 0.0 | 0.2 | GO:0060021 | palate development(GO:0060021) |
| 0.0 | 0.1 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.1 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.9 | GO:0097191 | extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.6 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.7 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 3.2 | GO:0032259 | methylation(GO:0032259) |
| 0.0 | 0.2 | GO:0007064 | mitotic sister chromatid cohesion(GO:0007064) |
| 0.0 | 0.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.7 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.5 | GO:0000281 | mitotic cytokinesis(GO:0000281) |
| 0.0 | 0.4 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.2 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.0 | 0.2 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.0 | 0.2 | GO:0048264 | determination of ventral identity(GO:0048264) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.2 | 0.7 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.2 | 1.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 0.8 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 0.5 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.1 | 0.5 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.3 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.4 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.1 | 2.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.1 | 0.5 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 0.6 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.1 | 0.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 1.4 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.4 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 3.0 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.8 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.1 | 0.7 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 0.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 1.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 0.7 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.3 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0000235 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.3 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.7 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.6 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.4 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.4 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 1.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle pole(GO:0097431) mitotic spindle midzone(GO:1990023) |
| 0.0 | 1.4 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.4 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.3 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.6 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.5 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 0.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.6 | GO:0005871 | kinesin complex(GO:0005871) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.3 | 1.0 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.2 | 2.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.2 | 1.4 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.2 | 0.6 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 2.9 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.1 | 1.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.6 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 0.7 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.6 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.1 | 0.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.8 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.1 | 0.4 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.4 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.1 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.4 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.1 | 0.5 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.2 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 1.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.1 | 0.5 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.4 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.1 | 0.4 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.2 | GO:0008061 | chitin binding(GO:0008061) |
| 0.1 | 2.1 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) RNA polymerase activity(GO:0034062) |
| 0.1 | 0.5 | GO:1904047 | S-adenosyl-L-methionine binding(GO:1904047) |
| 0.1 | 0.4 | GO:0001047 | core promoter sequence-specific DNA binding(GO:0001046) core promoter binding(GO:0001047) |
| 0.1 | 0.6 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.3 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.3 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.4 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.5 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 1.2 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.4 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.0 | 0.6 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.2 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 0.0 | 0.1 | GO:0043878 | aminobutyraldehyde dehydrogenase activity(GO:0019145) glyceraldehyde-3-phosphate dehydrogenase (NAD+) (non-phosphorylating) activity(GO:0043878) 4-trimethylammoniobutyraldehyde dehydrogenase activity(GO:0047105) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.0 | 0.2 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.0 | 0.6 | GO:0072542 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.0 | 0.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.4 | GO:0016866 | intramolecular transferase activity(GO:0016866) |
| 0.0 | 0.4 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.2 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.1 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.4 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.3 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.6 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.8 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.1 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.4 | GO:0042562 | hormone binding(GO:0042562) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 1.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.1 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 1.3 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.0 | 0.0 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 0.2 | GO:0004527 | exonuclease activity(GO:0004527) |
| 0.0 | 0.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.0 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 1.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 0.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.6 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.4 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.5 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 2.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.1 | 0.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.1 | 1.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.1 | 0.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.8 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.1 | 1.4 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 1.8 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.1 | 1.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 0.7 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.1 | 0.4 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 1.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.6 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 2.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.8 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.2 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.2 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.2 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |