Project
DANIO-CODE
Navigation
Downloads
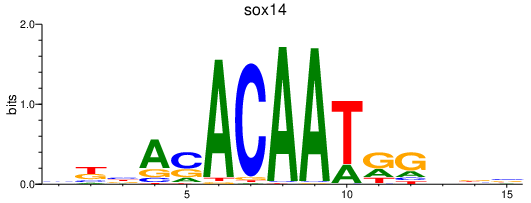
Results for sox14
Z-value: 1.10
Transcription factors associated with sox14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox14
|
ENSDARG00000070929 | SRY-box transcription factor 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox14 | dr10_dc_chr6_-_26569321_26569372 | 0.72 | 1.8e-03 | Click! |
Activity profile of sox14 motif
Sorted Z-values of sox14 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of sox14
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_12733209 | 2.74 |
ENSDART00000167021
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr11_-_29910947 | 2.35 |
ENSDART00000156121
|
scml2
|
sex comb on midleg-like 2 (Drosophila) |
| chr7_+_25642061 | 2.20 |
ENSDART00000135728
|
hmgb3a
|
high mobility group box 3a |
| chr1_+_25662652 | 2.10 |
ENSDART00000113020
|
tet2
|
tet methylcytosine dioxygenase 2 |
| chr16_+_26989962 | 2.09 |
ENSDART00000140673
|
galnt1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 |
| chr14_+_11151485 | 2.07 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr14_+_47326080 | 2.05 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr9_-_42894582 | 2.04 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr13_-_13163676 | 1.92 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr10_+_18994733 | 1.92 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr25_+_34069931 | 1.87 |
|
|
|
| chr19_-_7531709 | 1.79 |
ENSDART00000104750
|
mllt11
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 11 |
| chr23_+_36832325 | 1.76 |
|
|
|
| chr23_+_37754409 | 1.74 |
|
|
|
| chr11_+_15478585 | 1.73 |
ENSDART00000163289
ENSDART00000066033 |
gdf11
|
growth differentiation factor 11 |
| chr7_+_25641990 | 1.68 |
ENSDART00000129924
|
hmgb3a
|
high mobility group box 3a |
| chr6_+_7309023 | 1.67 |
ENSDART00000160128
|
erbb3a
|
erb-b2 receptor tyrosine kinase 3a |
| chr14_-_32449229 | 1.60 |
ENSDART00000172996
|
AL590151.1
|
ENSDARG00000053570 |
| chr10_+_33227967 | 1.60 |
ENSDART00000159666
|
myl10
|
myosin, light chain 10, regulatory |
| chr6_-_39767452 | 1.52 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr10_+_4962403 | 1.52 |
ENSDART00000134679
|
CU074419.2
|
ENSDARG00000093688 |
| chr9_-_11589126 | 1.51 |
ENSDART00000146832
|
cryba2b
|
crystallin, beta A2b |
| chr6_+_30681170 | 1.48 |
ENSDART00000112294
|
ttc22
|
tetratricopeptide repeat domain 22 |
| chr4_-_76620869 | 1.46 |
|
|
|
| chr23_-_19789409 | 1.43 |
ENSDART00000131860
|
rpl10
|
ribosomal protein L10 |
| chr10_-_27261937 | 1.43 |
|
|
|
| chr23_-_6707246 | 1.42 |
ENSDART00000023793
|
mipb
|
major intrinsic protein of lens fiber b |
| chr1_+_25662910 | 1.35 |
ENSDART00000113020
|
tet2
|
tet methylcytosine dioxygenase 2 |
| chr16_+_14697969 | 1.29 |
ENSDART00000133566
|
deptor
|
DEP domain containing MTOR-interacting protein |
| chr9_+_1177938 | 1.28 |
ENSDART00000176589
|
CABZ01066921.1
|
ENSDARG00000106418 |
| chr2_-_56224858 | 1.28 |
ENSDART00000060745
|
uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr13_-_31339870 | 1.25 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr8_-_13503775 | 1.24 |
ENSDART00000063834
|
ENSDARG00000043482
|
ENSDARG00000043482 |
| chr12_-_28840796 | 1.24 |
|
|
|
| chr13_-_13163801 | 1.24 |
ENSDART00000156968
|
fgfr3
|
fibroblast growth factor receptor 3 |
| chr21_+_20734431 | 1.21 |
ENSDART00000079732
|
oxct1b
|
3-oxoacid CoA transferase 1b |
| chr7_-_38590391 | 1.20 |
ENSDART00000037361
ENSDART00000173629 ENSDART00000173953 |
phf21aa
|
PHD finger protein 21Aa |
| chr4_-_72097569 | 1.19 |
ENSDART00000108669
|
zgc:171551
|
zgc:171551 |
| chr8_-_22945616 | 1.19 |
|
|
|
| chr10_+_5413893 | 1.18 |
ENSDART00000138821
|
nfil3
|
nuclear factor, interleukin 3 regulated |
| chr21_+_25651712 | 1.18 |
ENSDART00000135123
|
si:dkey-17e16.17
|
si:dkey-17e16.17 |
| chr21_-_35806638 | 1.17 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr10_-_11806373 | 1.16 |
ENSDART00000009715
|
adamts6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr4_+_28860191 | 1.16 |
ENSDART00000110358
ENSDART00000162069 |
znf1059
|
zinc finger protein 1059 |
| chr3_-_38642067 | 1.12 |
ENSDART00000155518
|
znf281a
|
zinc finger protein 281a |
| chr6_+_33556031 | 1.11 |
ENSDART00000147625
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr23_+_36032206 | 1.11 |
ENSDART00000103132
|
hoxc4a
|
homeobox C4a |
| chr8_-_49183600 | 1.11 |
ENSDART00000134269
|
fkbp1aa
|
FK506 binding protein 1Aa |
| chr12_+_29931452 | 1.10 |
ENSDART00000153394
|
ablim1b
|
actin binding LIM protein 1b |
| chr19_-_19290260 | 1.09 |
|
|
|
| chr2_+_56339323 | 1.09 |
ENSDART00000123392
|
rab11bb
|
RAB11B, member RAS oncogene family, b |
| chr13_-_31304614 | 1.07 |
|
|
|
| chr25_-_28691412 | 1.05 |
ENSDART00000067066
|
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
| chr14_-_25301744 | 1.04 |
ENSDART00000131886
|
slc25a48
|
solute carrier family 25, member 48 |
| chr21_+_43674754 | 1.03 |
ENSDART00000136025
|
tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr18_-_13234871 | 1.00 |
ENSDART00000142942
|
klhl36
|
kelch-like family member 36 |
| chr9_-_306569 | 0.99 |
ENSDART00000166059
|
si:ch211-166e11.5
|
si:ch211-166e11.5 |
| chr9_-_23340795 | 0.97 |
ENSDART00000133017
|
lypd6b
|
LY6/PLAUR domain containing 6B |
| chr12_-_17357133 | 0.97 |
ENSDART00000079115
|
papss2b
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2b |
| chr8_-_17031599 | 0.97 |
ENSDART00000132687
|
rab3c
|
RAB3C, member RAS oncogene family |
| chr19_+_47719438 | 0.96 |
ENSDART00000138295
|
ext1c
|
exostoses (multiple) 1c |
| chr9_+_1177862 | 0.94 |
ENSDART00000176589
|
CABZ01066921.1
|
ENSDARG00000106418 |
| chr11_-_18393802 | 0.93 |
ENSDART00000125453
|
dido1
|
death inducer-obliterator 1 |
| chr19_-_44043806 | 0.93 |
ENSDART00000138009
ENSDART00000086138 |
fam83hb
|
family with sequence similarity 83, member Hb |
| chr9_-_22371512 | 0.92 |
ENSDART00000101809
|
crygm2d6
|
crystallin, gamma M2d6 |
| chr23_-_7892862 | 0.90 |
ENSDART00000157612
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr7_-_57030751 | 0.89 |
|
|
|
| chr12_+_22558928 | 0.88 |
ENSDART00000077548
|
ablim2
|
actin binding LIM protein family, member 2 |
| chr25_-_36760720 | 0.87 |
ENSDART00000111862
|
ldlrad3
|
low density lipoprotein receptor class A domain containing 3 |
| chr15_+_15479719 | 0.85 |
ENSDART00000127436
|
zgc:92630
|
zgc:92630 |
| chr13_-_20387917 | 0.84 |
ENSDART00000167916
ENSDART00000165310 ENSDART00000168955 |
gfra1a
|
gdnf family receptor alpha 1a |
| chr15_-_33960811 | 0.82 |
ENSDART00000158325
|
n4bp2l2
|
NEDD4 binding protein 2-like 2 |
| chr2_+_36957471 | 0.82 |
ENSDART00000143915
|
gadd45bb
|
growth arrest and DNA-damage-inducible, beta b |
| chr18_+_21133694 | 0.82 |
ENSDART00000060015
|
chka
|
choline kinase alpha |
| chr22_+_5657904 | 0.80 |
ENSDART00000138102
|
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr1_-_32825184 | 0.79 |
ENSDART00000075632
|
creb1a
|
cAMP responsive element binding protein 1a |
| chr11_-_42626842 | 0.79 |
ENSDART00000052912
|
pcdh20
|
protocadherin 20 |
| chr22_+_38837247 | 0.77 |
ENSDART00000144318
|
anos1b
|
anosmin 1b |
| chr18_+_37034218 | 0.75 |
|
|
|
| chr19_-_44043682 | 0.75 |
ENSDART00000138009
ENSDART00000086138 |
fam83hb
|
family with sequence similarity 83, member Hb |
| chr15_+_657554 | 0.72 |
ENSDART00000156007
|
si:ch73-144d13.8
|
si:ch73-144d13.8 |
| chr11_+_23694891 | 0.70 |
ENSDART00000000486
|
cntn2
|
contactin 2 |
| chr19_-_41819752 | 0.69 |
ENSDART00000167772
|
shfm1
|
split hand/foot malformation (ectrodactyly) type 1 |
| chr2_-_39052185 | 0.68 |
ENSDART00000048838
|
rbp2b
|
retinol binding protein 2b, cellular |
| chr17_-_5703094 | 0.68 |
ENSDART00000058894
|
si:ch73-340m8.2
|
si:ch73-340m8.2 |
| chr3_+_52901602 | 0.67 |
ENSDART00000114343
|
brd4
|
bromodomain containing 4 |
| chr6_+_33556155 | 0.66 |
ENSDART00000147625
|
pik3r3b
|
phosphoinositide-3-kinase, regulatory subunit 3b (gamma) |
| chr4_-_25807510 | 0.65 |
ENSDART00000122881
|
tmcc3
|
transmembrane and coiled-coil domain family 3 |
| chr23_-_17078715 | 0.64 |
ENSDART00000080545
|
dnmt3bb.2
|
DNA (cytosine-5-)-methyltransferase 3 beta, duplicate b.2 |
| chr4_-_38406131 | 0.63 |
|
|
|
| chr5_-_57016269 | 0.62 |
ENSDART00000074264
|
crfb12
|
cytokine receptor family member B12 |
| chr18_+_45203251 | 0.61 |
ENSDART00000015786
|
gyltl1b
|
glycosyltransferase-like 1b |
| chr12_+_22455344 | 0.61 |
ENSDART00000172053
|
capgb
|
capping protein (actin filament), gelsolin-like b |
| chr6_-_37765778 | 0.61 |
ENSDART00000149068
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr16_-_42157129 | 0.60 |
|
|
|
| chr14_-_952510 | 0.59 |
ENSDART00000130801
|
acsl1b
|
acyl-CoA synthetase long-chain family member 1b |
| chr6_+_43429115 | 0.57 |
ENSDART00000056457
|
mitfa
|
microphthalmia-associated transcription factor a |
| chr21_-_21477423 | 0.57 |
ENSDART00000164076
|
pvrl3b
|
poliovirus receptor-related 3b |
| chr8_+_26502909 | 0.56 |
ENSDART00000046863
|
sema3bl
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3bl |
| chr23_-_3815492 | 0.56 |
ENSDART00000131536
|
hmga1a
|
high mobility group AT-hook 1a |
| chr19_-_7531649 | 0.56 |
ENSDART00000104750
|
mllt11
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 11 |
| chr16_+_26990064 | 0.56 |
ENSDART00000140673
|
galnt1
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 |
| chr15_-_37832225 | 0.54 |
ENSDART00000059568
|
si:ch211-137j23.6
|
si:ch211-137j23.6 |
| chr19_-_34328673 | 0.54 |
ENSDART00000164563
|
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr21_-_2222412 | 0.53 |
ENSDART00000169448
|
si:ch73-299h12.1
|
si:ch73-299h12.1 |
| chr5_+_296856 | 0.53 |
|
|
|
| chr14_-_29565869 | 0.50 |
ENSDART00000172349
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr20_+_35479428 | 0.50 |
ENSDART00000159483
ENSDART00000031091 |
BX511024.1
vsnl1a
|
ENSDARG00000104812 visinin-like 1a |
| chr1_+_2074153 | 0.48 |
ENSDART00000058877
|
rap2ab
|
RAP2A, member of RAS oncogene family b |
| chr13_+_12651778 | 0.47 |
ENSDART00000090000
|
si:ch211-233a24.2
|
si:ch211-233a24.2 |
| chr5_-_55745399 | 0.47 |
|
|
|
| chr12_-_425381 | 0.46 |
ENSDART00000083827
|
hs3st3l
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 3-like |
| chr20_+_49975268 | 0.46 |
ENSDART00000008808
|
hmbox1b
|
homeobox containing 1 b |
| chr20_+_53716378 | 0.46 |
ENSDART00000139794
|
pak6b
|
p21 protein (Cdc42/Rac)-activated kinase 6b |
| chr22_+_18762028 | 0.46 |
ENSDART00000105399
|
cbarpb
|
calcium channel, voltage-dependent, beta subunit associated regulatory protein b |
| chr20_+_54247734 | 0.45 |
ENSDART00000143172
|
wdr20b
|
WD repeat domain 20b |
| chr16_+_25158949 | 0.44 |
ENSDART00000155465
|
si:dkeyp-84f3.9
|
si:dkeyp-84f3.9 |
| chr6_+_30469823 | 0.44 |
ENSDART00000121492
|
FP236735.1
|
ENSDARG00000087805 |
| chr13_-_13163647 | 0.43 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr4_-_13394059 | 0.43 |
ENSDART00000175909
ENSDART00000174861 |
BX511038.1
|
ENSDARG00000108089 |
| chr3_-_28534446 | 0.43 |
ENSDART00000151670
|
fbxl16
|
F-box and leucine-rich repeat protein 16 |
| chr23_+_42920817 | 0.42 |
ENSDART00000055577
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr25_-_22850980 | 0.42 |
ENSDART00000175228
|
CU462979.1
|
ENSDARG00000107627 |
| chr17_-_37266525 | 0.42 |
ENSDART00000128715
|
kif3cb
|
kinesin family member 3Cb |
| chr11_-_18016658 | 0.41 |
|
|
|
| chr9_-_13991784 | 0.41 |
ENSDART00000061156
|
raph1a
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1a |
| chr13_-_31340029 | 0.40 |
ENSDART00000076574
|
rtn1a
|
reticulon 1a |
| chr3_-_49104540 | 0.40 |
ENSDART00000138503
|
btr02
|
bloodthirsty-related gene family, member 2 |
| chr25_-_34607624 | 0.40 |
ENSDART00000128017
|
CU302436.1
|
ENSDARG00000051726 |
| chr2_-_26987186 | 0.40 |
ENSDART00000132854
|
u2surp
|
U2 snRNP-associated SURP domain containing |
| chr7_+_54347388 | 0.39 |
ENSDART00000158898
|
fgf4
|
fibroblast growth factor 4 |
| chr11_+_13000957 | 0.38 |
ENSDART00000161532
|
zfyve9b
|
zinc finger, FYVE domain containing 9b |
| chr22_-_15666856 | 0.38 |
ENSDART00000141861
|
si:ch1073-396h14.1
|
si:ch1073-396h14.1 |
| chr25_+_21735567 | 0.37 |
ENSDART00000148299
|
ckmt1
|
creatine kinase, mitochondrial 1 |
| chr5_-_24208617 | 0.35 |
ENSDART00000033630
|
ENSDARG00000022772
|
ENSDARG00000022772 |
| chr6_+_3356712 | 0.35 |
ENSDART00000151607
|
faah
|
fatty acid amide hydrolase |
| chr10_-_27262050 | 0.34 |
|
|
|
| chr13_-_20388016 | 0.34 |
ENSDART00000167916
ENSDART00000165310 ENSDART00000168955 |
gfra1a
|
gdnf family receptor alpha 1a |
| chr14_+_29933552 | 0.34 |
|
|
|
| chr1_-_32825274 | 0.34 |
ENSDART00000075632
|
creb1a
|
cAMP responsive element binding protein 1a |
| chr3_-_25144722 | 0.34 |
ENSDART00000055445
|
ddx5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr8_-_23754376 | 0.32 |
ENSDART00000114800
|
INAVA
|
innate immunity activator |
| chr21_-_22694225 | 0.32 |
ENSDART00000101797
|
fbxo40.1
|
F-box protein 40, tandem duplicate 1 |
| chr19_-_28382815 | 0.31 |
ENSDART00000110016
|
ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr19_+_47719402 | 0.30 |
ENSDART00000138295
|
ext1c
|
exostoses (multiple) 1c |
| chr10_-_175160 | 0.30 |
|
|
|
| chr10_-_9294185 | 0.30 |
|
|
|
| chr4_-_22509152 | 0.29 |
ENSDART00000176950
|
ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr14_+_23419894 | 0.29 |
ENSDART00000006373
|
ndfip1
|
Nedd4 family interacting protein 1 |
| chr23_-_27930478 | 0.29 |
ENSDART00000043462
|
acvrl1
|
activin A receptor type II-like 1 |
| chr12_+_2958777 | 0.29 |
ENSDART00000082297
|
lrrc45
|
leucine rich repeat containing 45 |
| chr13_+_5442296 | 0.28 |
ENSDART00000134506
|
si:dkey-196j8.2
|
si:dkey-196j8.2 |
| chr11_+_11284217 | 0.28 |
ENSDART00000026814
|
ptp4a1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr12_-_2766066 | 0.27 |
ENSDART00000152682
|
ubtd1b
|
ubiquitin domain containing 1b |
| chr16_-_45450439 | 0.27 |
|
|
|
| chr20_-_21775455 | 0.27 |
|
|
|
| chr17_+_33862300 | 0.27 |
|
|
|
| chr16_+_54350976 | 0.27 |
ENSDART00000172622
ENSDART00000020033 |
xrcc1
|
X-ray repair complementing defective repair in Chinese hamster cells 1 |
| chr21_+_37751640 | 0.26 |
ENSDART00000076328
|
pgrmc1
|
progesterone receptor membrane component 1 |
| chr12_-_27497159 | 0.25 |
ENSDART00000066282
|
dhx8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr14_-_2268924 | 0.24 |
ENSDART00000160205
|
pcdh2ab12
|
protocadherin 2 alpha b 12 |
| chr16_-_25793780 | 0.23 |
|
|
|
| chr10_-_27262098 | 0.22 |
|
|
|
| chr4_-_47142181 | 0.21 |
ENSDART00000150426
|
znf1064
|
zinc finger protein 1064 |
| chr23_-_12305794 | 0.20 |
|
|
|
| chr7_+_28782951 | 0.19 |
|
|
|
| chr12_-_16035939 | 0.18 |
ENSDART00000090881
|
si:dkey-100n10.2
|
si:dkey-100n10.2 |
| chr19_+_46631965 | 0.18 |
ENSDART00000158703
|
vps28
|
vacuolar protein sorting 28 (yeast) |
| chr10_-_14604287 | 0.18 |
|
|
|
| chr25_+_3551924 | 0.17 |
ENSDART00000154348
|
prnprs3
|
prion protein, related sequence 3 |
| chr17_+_8768744 | 0.17 |
|
|
|
| chr19_-_44044039 | 0.17 |
ENSDART00000138009
ENSDART00000086138 |
fam83hb
|
family with sequence similarity 83, member Hb |
| chr7_-_38590218 | 0.17 |
ENSDART00000133491
|
phf21aa
|
PHD finger protein 21Aa |
| chr1_-_499082 | 0.16 |
ENSDART00000144406
ENSDART00000031635 |
ercc5
|
excision repair cross-complementation group 5 |
| chr8_+_18800414 | 0.16 |
ENSDART00000160732
|
mpnd
|
MPN domain containing |
| chr2_-_56265681 | 0.16 |
ENSDART00000036240
|
cers4b
|
ceramide synthase 4b |
| chr5_+_38143180 | 0.15 |
ENSDART00000137658
|
si:dkey-58f10.6
|
si:dkey-58f10.6 |
| chr19_+_42768795 | 0.14 |
|
|
|
| chr18_+_15872911 | 0.14 |
ENSDART00000141800
|
eea1
|
early endosome antigen 1 |
| chr5_-_21430034 | 0.14 |
|
|
|
| chr18_+_37279031 | 0.13 |
ENSDART00000135444
|
tbcb
|
tubulin folding cofactor B |
| chr19_-_15324958 | 0.13 |
ENSDART00000055619
|
phactr4a
|
phosphatase and actin regulator 4a |
| chr18_+_14509074 | 0.13 |
ENSDART00000146472
|
kcng4a
|
potassium voltage-gated channel, subfamily G, member 4a |
| chr5_+_38904048 | 0.12 |
ENSDART00000121460
|
prdm8b
|
PR domain containing 8b |
| chr13_-_49430733 | 0.11 |
|
|
|
| chr6_-_39767422 | 0.11 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr19_+_13492428 | 0.10 |
ENSDART00000165033
|
srsf4
|
serine/arginine-rich splicing factor 4 |
| chr3_+_41573049 | 0.09 |
ENSDART00000155440
|
eif3ba
|
eukaryotic translation initiation factor 3, subunit Ba |
| chr7_+_55217611 | 0.09 |
ENSDART00000149915
|
cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr8_-_45752084 | 0.08 |
ENSDART00000025620
|
ppiaa
|
peptidylprolyl isomerase Aa (cyclophilin A) |
| chr2_-_37151666 | 0.08 |
ENSDART00000146123
|
elavl1
|
ELAV like RNA binding protein 1 |
| chr23_-_27930393 | 0.08 |
ENSDART00000043462
|
acvrl1
|
activin A receptor type II-like 1 |
| chr20_+_34019004 | 0.08 |
ENSDART00000161363
|
lmx1a
|
LIM homeobox transcription factor 1, alpha |
| chr1_+_53353287 | 0.07 |
ENSDART00000012104
|
dla
|
deltaA |
| chr24_-_31257591 | 0.07 |
ENSDART00000158808
|
cnn3a
|
calponin 3, acidic a |
| chr13_+_23767450 | 0.07 |
ENSDART00000002244
|
sf3b5
|
splicing factor 3b, subunit 5 |
| chr6_+_9801649 | 0.06 |
ENSDART00000149537
|
sucla2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr5_+_38143502 | 0.05 |
ENSDART00000137658
|
si:dkey-58f10.6
|
si:dkey-58f10.6 |
| chr2_+_37992682 | 0.05 |
ENSDART00000179233
|
CR376839.1
|
ENSDARG00000108815 |
| chr15_-_723851 | 0.05 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.5 | GO:0006211 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.5 | 2.3 | GO:0090199 | regulation of mitochondrial membrane potential(GO:0051881) membrane depolarization(GO:0051899) regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.3 | 1.2 | GO:0046950 | cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.2 | 1.6 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.2 | 2.7 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) mitotic DNA replication(GO:1902969) |
| 0.2 | 0.9 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.2 | 1.4 | GO:0006833 | water transport(GO:0006833) |
| 0.2 | 0.7 | GO:0048682 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.2 | 1.9 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.2 | 1.0 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 0.7 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.2 | 1.0 | GO:0034036 | sulfate assimilation(GO:0000103) purine ribonucleoside bisphosphate metabolic process(GO:0034035) purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.2 | 0.5 | GO:0051701 | interaction with host(GO:0051701) |
| 0.1 | 0.3 | GO:1903429 | regulation of cell maturation(GO:1903429) |
| 0.1 | 0.5 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 1.8 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.9 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 3.6 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 0.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.6 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.1 | 0.5 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.1 | 1.4 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.2 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.1 | 4.5 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 0.4 | GO:0034405 | response to fluid shear stress(GO:0034405) |
| 0.1 | 1.0 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 1.0 | GO:0051181 | cofactor transport(GO:0051181) |
| 0.1 | 0.8 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.1 | 0.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.7 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.0 | 0.4 | GO:0006599 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.0 | 0.3 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 1.2 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.8 | GO:0048922 | posterior lateral line neuromast deposition(GO:0048922) |
| 0.0 | 1.3 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.9 | GO:0030032 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 1.1 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 1.0 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.6 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.5 | GO:0002429 | immune response-activating cell surface receptor signaling pathway(GO:0002429) |
| 0.0 | 0.1 | GO:0055016 | hypochord development(GO:0055016) |
| 0.0 | 1.8 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.5 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.0 | 3.1 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.9 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.6 | GO:0035338 | long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.2 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.0 | 0.4 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 1.9 | GO:0008284 | positive regulation of cell proliferation(GO:0008284) |
| 0.0 | 0.6 | GO:0007599 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.0 | 0.2 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 2.6 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.4 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.6 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 1.1 | GO:0045055 | regulated exocytosis(GO:0045055) |
| 0.0 | 0.1 | GO:0003313 | heart rudiment development(GO:0003313) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 1.6 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 1.7 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.1 | 0.9 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 2.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.4 | GO:0070724 | BMP receptor complex(GO:0070724) |
| 0.1 | 0.9 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 1.7 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 1.2 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 1.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.3 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 1.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0042709 | succinate-CoA ligase complex(GO:0042709) |
| 0.0 | 1.6 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.5 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.7 | 2.7 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.3 | 1.0 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.3 | 1.3 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.3 | 1.2 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.3 | 1.9 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.2 | 1.4 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 1.6 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.2 | 3.9 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 3.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 1.0 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.2 | 4.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.8 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 1.0 | GO:0015101 | organic cation transmembrane transporter activity(GO:0015101) |
| 0.1 | 0.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 1.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 0.3 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.1 | 0.7 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.3 | GO:0031386 | protein tag(GO:0031386) |
| 0.1 | 0.6 | GO:0047676 | arachidonate-CoA ligase activity(GO:0047676) |
| 0.1 | 1.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.4 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.0 | 0.9 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 1.4 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.6 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 2.0 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.7 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 1.2 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.0 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) succinate-CoA ligase (GDP-forming) activity(GO:0004776) |
| 0.0 | 2.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 1.7 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.5 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.6 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.5 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.0 | 0.2 | GO:0070122 | isopeptidase activity(GO:0070122) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 1.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.7 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.2 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.3 | 1.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 2.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 0.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.5 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 1.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |