Project
DANIO-CODE
Navigation
Downloads
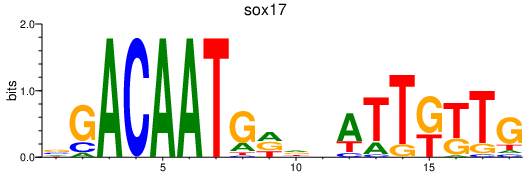
Results for sox17
Z-value: 0.59
Transcription factors associated with sox17
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox17
|
ENSDARG00000101717 | SRY-box transcription factor 17 |
Activity profile of sox17 motif
Sorted Z-values of sox17 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of sox17
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_20571619 | 2.33 |
ENSDART00000040074
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr23_+_32174669 | 1.33 |
ENSDART00000000992
|
ENSDARG00000000887
|
ENSDARG00000000887 |
| chr22_-_20236308 | 1.23 |
|
|
|
| chr5_-_23211957 | 1.18 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr12_-_3042394 | 1.12 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr5_-_32433490 | 1.08 |
|
|
|
| chr24_-_9880951 | 1.06 |
ENSDART00000137250
|
zgc:173856
|
zgc:173856 |
| chr14_+_29932533 | 1.05 |
ENSDART00000017122
|
asah1a
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1a |
| chr21_+_3627192 | 1.03 |
ENSDART00000139813
|
tor1
|
torsin family 1 |
| chr5_-_49417365 | 1.00 |
|
|
|
| chr22_+_1837102 | 0.99 |
ENSDART00000163288
|
znf1174
|
zinc finger protein 1174 |
| chr21_-_32747592 | 0.97 |
|
|
|
| chr24_+_9887575 | 0.96 |
ENSDART00000172773
|
si:ch211-146l10.8
|
si:ch211-146l10.8 |
| chr13_+_11418060 | 0.95 |
ENSDART00000166908
|
desi2
|
desumoylating isopeptidase 2 |
| chr18_+_17545564 | 0.94 |
ENSDART00000061007
|
mt2
|
metallothionein 2 |
| chr7_+_66660882 | 0.91 |
ENSDART00000082664
|
sbf2
|
SET binding factor 2 |
| chr19_+_33360759 | 0.89 |
ENSDART00000052091
|
atp6v1c1b
|
ATPase, H+ transporting, lysosomal, V1 subunit C1b |
| chr7_+_38078338 | 0.86 |
ENSDART00000010046
|
rhpn2
|
rhophilin, Rho GTPase binding protein 2 |
| chr22_+_30041252 | 0.78 |
ENSDART00000165313
|
si:dkey-286j15.3
|
si:dkey-286j15.3 |
| chr18_-_958923 | 0.78 |
|
|
|
| chr23_+_19728953 | 0.78 |
ENSDART00000104441
|
abhd6b
|
abhydrolase domain containing 6b |
| chr17_-_16414577 | 0.78 |
ENSDART00000150149
|
tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr2_-_26941084 | 0.76 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr3_+_13034056 | 0.76 |
|
|
|
| KN150339v1_-_39357 | 0.74 |
ENSDART00000169638
|
wu:fa19b12
|
wu:fa19b12 |
| chr3_+_18287783 | 0.73 |
ENSDART00000144690
|
tbc1d16
|
TBC1 domain family, member 16 |
| chr13_+_11417882 | 0.70 |
ENSDART00000034935
|
desi2
|
desumoylating isopeptidase 2 |
| chr5_-_32433573 | 0.69 |
|
|
|
| chr8_-_44617367 | 0.68 |
ENSDART00000063396
|
bag4
|
BCL2-associated athanogene 4 |
| chr2_-_26941232 | 0.68 |
ENSDART00000134685
ENSDART00000056787 |
zgc:113691
|
zgc:113691 |
| chr10_-_45182378 | 0.68 |
ENSDART00000161815
|
polm
|
polymerase (DNA directed), mu |
| chr13_-_402878 | 0.68 |
ENSDART00000038315
|
nvl
|
nuclear VCP-like |
| chr23_+_30803518 | 0.68 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr1_+_8468971 | 0.68 |
ENSDART00000081343
|
plk1
|
polo-like kinase 1 (Drosophila) |
| chr3_+_18287645 | 0.68 |
ENSDART00000136243
|
tbc1d16
|
TBC1 domain family, member 16 |
| chr10_+_19625897 | 0.67 |
|
|
|
| chr16_+_25192196 | 0.66 |
ENSDART00000033211
|
zgc:66448
|
zgc:66448 |
| chr1_-_45958767 | 0.66 |
ENSDART00000053222
|
rcbtb1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
| chr13_+_50990946 | 0.65 |
|
|
|
| chr2_+_223139 | 0.65 |
ENSDART00000113021
|
dhx30
|
DEAH (Asp-Glu-Ala-His) box helicase 30 |
| chr20_-_21033044 | 0.65 |
ENSDART00000152415
|
btbd6b
|
BTB (POZ) domain containing 6b |
| chr18_+_20571460 | 0.64 |
ENSDART00000151990
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr21_-_37750338 | 0.63 |
ENSDART00000133585
|
fam114a2
|
family with sequence similarity 114, member A2 |
| chr21_+_11943637 | 0.63 |
ENSDART00000141306
|
zgc:162344
|
zgc:162344 |
| chr14_+_15788442 | 0.62 |
ENSDART00000159181
|
immt
|
inner membrane protein, mitochondrial (mitofilin) |
| chr7_+_10319999 | 0.61 |
ENSDART00000168801
|
zfand6
|
zinc finger, AN1-type domain 6 |
| chr1_+_52686620 | 0.61 |
ENSDART00000176087
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr19_-_43434166 | 0.61 |
|
|
|
| chr12_+_35855929 | 0.61 |
|
|
|
| chr1_+_52686770 | 0.60 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr9_-_21287478 | 0.60 |
ENSDART00000018570
|
wars2
|
tryptophanyl tRNA synthetase 2, mitochondrial |
| KN149908v1_+_8072 | 0.60 |
|
|
|
| chr12_-_30244042 | 0.59 |
ENSDART00000152878
|
tdrd1
|
tudor domain containing 1 |
| chr2_+_38042821 | 0.58 |
ENSDART00000134211
ENSDART00000144868 |
hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr18_+_35253031 | 0.58 |
|
|
|
| chr13_-_33186642 | 0.57 |
ENSDART00000110295
|
tmem234
|
transmembrane protein 234 |
| chr24_+_12691540 | 0.57 |
|
|
|
| chr21_+_3627046 | 0.57 |
ENSDART00000139813
|
tor1
|
torsin family 1 |
| chr22_+_1335355 | 0.57 |
ENSDART00000169629
|
znf45l
|
zinc finger 45 like |
| chr12_-_926596 | 0.56 |
ENSDART00000088351
|
SPAG9 (1 of many)
|
sperm associated antigen 9 |
| chr8_-_44617324 | 0.55 |
ENSDART00000063396
|
bag4
|
BCL2-associated athanogene 4 |
| chr12_-_30244575 | 0.55 |
ENSDART00000152981
|
tdrd1
|
tudor domain containing 1 |
| chr5_+_51186708 | 0.55 |
ENSDART00000165276
|
papd4
|
PAP associated domain containing 4 |
| chr16_-_55211053 | 0.55 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr24_-_16835352 | 0.55 |
ENSDART00000005331
ENSDART00000128446 |
klhl15
|
kelch-like family member 15 |
| chr15_-_18273640 | 0.54 |
ENSDART00000141508
|
btr16
|
bloodthirsty-related gene family, member 16 |
| chr23_+_32175042 | 0.54 |
ENSDART00000138849
|
ENSDARG00000000887
|
ENSDARG00000000887 |
| chr19_-_17306473 | 0.54 |
ENSDART00000007906
|
stmn1a
|
stathmin 1a |
| chr22_-_9790536 | 0.54 |
|
|
|
| chr12_-_49212400 | 0.52 |
ENSDART00000112479
|
acadsb
|
acyl-CoA dehydrogenase, short/branched chain |
| chr12_-_33256599 | 0.52 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr22_+_32274436 | 0.51 |
ENSDART00000092082
|
manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr5_+_38498787 | 0.51 |
ENSDART00000006079
|
bmp2k
|
BMP2 inducible kinase |
| chr20_-_46079578 | 0.50 |
ENSDART00000153228
|
AL929237.3
|
ENSDARG00000096676 |
| chr8_-_44617200 | 0.50 |
ENSDART00000063396
|
bag4
|
BCL2-associated athanogene 4 |
| chr7_+_10320142 | 0.49 |
ENSDART00000168801
|
zfand6
|
zinc finger, AN1-type domain 6 |
| chr5_+_36904127 | 0.49 |
ENSDART00000165465
|
si:ch1073-224n8.1
|
si:ch1073-224n8.1 |
| chr3_+_28729443 | 0.48 |
ENSDART00000077235
|
alg1
|
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr5_+_35930704 | 0.48 |
ENSDART00000006899
|
igbp1
|
immunoglobulin (CD79A) binding protein 1 |
| chr23_-_24615681 | 0.48 |
ENSDART00000132265
|
atp13a2
|
ATPase type 13A2 |
| chr15_+_6462943 | 0.48 |
ENSDART00000065824
|
bace2
|
beta-site APP-cleaving enzyme 2 |
| chr11_-_27290434 | 0.48 |
ENSDART00000173444
|
ptpdc1a
|
protein tyrosine phosphatase domain containing 1a |
| chr25_-_13394261 | 0.48 |
ENSDART00000056721
|
ldhd
|
lactate dehydrogenase D |
| chr16_+_35952050 | 0.48 |
ENSDART00000162411
|
sh3d21
|
SH3 domain containing 21 |
| chr18_+_20571513 | 0.47 |
ENSDART00000136710
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr20_-_49067167 | 0.47 |
ENSDART00000163071
ENSDART00000170617 |
xrn2
|
5'-3' exoribonuclease 2 |
| chr13_-_4535689 | 0.47 |
ENSDART00000023803
|
c1d
|
C1D nuclear receptor corepressor |
| chr15_-_31104074 | 0.46 |
ENSDART00000157005
|
nf1a
|
neurofibromin 1a |
| chr2_+_22753634 | 0.46 |
ENSDART00000171853
|
LMO4
|
zgc:56628 |
| chr13_-_31557441 | 0.45 |
ENSDART00000085989
|
trmt5
|
TRM5 tRNA methyltransferase 5 homolog (S. cerevisiae) |
| chr25_+_20596490 | 0.45 |
ENSDART00000073648
|
kxd1
|
KxDL motif containing 1 |
| chr18_-_46185361 | 0.45 |
ENSDART00000021192
|
kcnk6
|
potassium channel, subfamily K, member 6 |
| chr24_-_10935904 | 0.45 |
ENSDART00000003195
|
chmp4c
|
charged multivesicular body protein 4C |
| chr11_+_25302044 | 0.45 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr20_+_40560443 | 0.45 |
ENSDART00000153119
|
serinc1
|
serine incorporator 1 |
| chr16_+_41767185 | 0.44 |
ENSDART00000015170
|
mtdhb
|
metadherin b |
| chr23_+_30803887 | 0.44 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr4_+_13413303 | 0.44 |
ENSDART00000003694
|
cand1
|
cullin-associated and neddylation-dissociated 1 |
| chr13_+_11307037 | 0.44 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr5_+_51186666 | 0.44 |
ENSDART00000165276
|
papd4
|
PAP associated domain containing 4 |
| chr1_-_51863300 | 0.44 |
ENSDART00000004233
|
acy3.2
|
aspartoacylase (aminocyclase) 3, tandem duplicate 2 |
| chr2_+_38042740 | 0.43 |
ENSDART00000134211
ENSDART00000144868 |
hnrnpc
|
heterogeneous nuclear ribonucleoprotein C |
| chr23_+_28464642 | 0.43 |
ENSDART00000133736
|
CT025585.1
|
ENSDARG00000093306 |
| chr14_+_26268810 | 0.43 |
ENSDART00000173056
|
arap3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr21_+_11943609 | 0.43 |
ENSDART00000155426
ENSDART00000102463 |
zgc:162344
|
zgc:162344 |
| chr17_-_11904068 | 0.43 |
ENSDART00000105236
|
smyd3
|
SET and MYND domain containing 3 |
| chr12_-_3042573 | 0.43 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr23_+_16848278 | 0.42 |
ENSDART00000104791
|
zgc:153722
|
zgc:153722 |
| chr20_-_46079529 | 0.42 |
ENSDART00000153228
|
AL929237.3
|
ENSDARG00000096676 |
| chr6_-_8156471 | 0.42 |
ENSDART00000081561
|
ilf3a
|
interleukin enhancer binding factor 3a |
| chr11_-_33605979 | 0.42 |
ENSDART00000171439
|
si:ch211-227n13.3
|
si:ch211-227n13.3 |
| chr3_+_34690574 | 0.41 |
|
|
|
| chr24_-_24884787 | 0.41 |
ENSDART00000168254
|
nup58
|
nucleoporin 58 |
| chr4_-_3215313 | 0.41 |
ENSDART00000112210
|
plekha5
|
pleckstrin homology domain containing, family A member 5 |
| chr7_-_39107780 | 0.41 |
ENSDART00000173659
|
slc8b1
|
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
| chr17_-_19943767 | 0.41 |
ENSDART00000154251
|
chrm3a
|
cholinergic receptor, muscarinic 3a |
| chr1_+_57891538 | 0.41 |
ENSDART00000158425
|
FO704618.1
|
ENSDARG00000105176 |
| chr11_+_25302180 | 0.41 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr6_-_29386437 | 0.41 |
ENSDART00000168905
|
tmem131
|
transmembrane protein 131 |
| chr7_-_13555920 | 0.40 |
ENSDART00000172852
|
si:cabz01068815.1
|
si:cabz01068815.1 |
| chr13_+_31557497 | 0.40 |
ENSDART00000076479
|
slc38a6
|
solute carrier family 38, member 6 |
| chr21_+_7570420 | 0.40 |
ENSDART00000161921
|
zgc:113019
|
zgc:113019 |
| chr13_-_30938624 | 0.40 |
ENSDART00000112653
|
wdfy4
|
WDFY family member 4 |
| chr9_-_46475965 | 0.40 |
ENSDART00000165238
|
hdac4
|
histone deacetylase 4 |
| chr19_+_43316957 | 0.39 |
ENSDART00000151298
|
arpp21
|
cAMP-regulated phosphoprotein, 21 |
| chr25_+_20596613 | 0.39 |
ENSDART00000073648
|
kxd1
|
KxDL motif containing 1 |
| chr16_-_28723759 | 0.39 |
ENSDART00000148456
|
abcb4
|
ATP-binding cassette, sub-family B (MDR/TAP), member 4 |
| chr12_-_16403282 | 0.38 |
ENSDART00000037601
|
rpp30
|
ribonuclease P/MRP 30 subunit |
| chr15_-_31103957 | 0.38 |
ENSDART00000080328
|
nf1a
|
neurofibromin 1a |
| chr12_-_3042545 | 0.38 |
ENSDART00000002867
ENSDART00000126315 |
rfng
|
RFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr1_+_57878303 | 0.38 |
ENSDART00000162460
|
si:ch73-221f6.3
|
si:ch73-221f6.3 |
| chr18_-_44365869 | 0.37 |
ENSDART00000166935
|
prdm10
|
PR domain containing 10 |
| chr16_-_47366405 | 0.37 |
ENSDART00000169697
|
mios
|
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr18_-_21736549 | 0.37 |
ENSDART00000089853
|
fan1
|
FANCD2/FANCI-associated nuclease 1 |
| chr17_-_20198477 | 0.37 |
ENSDART00000063523
|
mgmt
|
O-6-methylguanine-DNA methyltransferase |
| chr16_+_28829383 | 0.37 |
ENSDART00000122433
|
trim33l
|
tripartite motif containing 33, like |
| chr9_+_38648006 | 0.37 |
ENSDART00000142512
|
mcm3ap
|
minichromosome maintenance complex component 3 associated protein |
| chr10_-_6587281 | 0.36 |
ENSDART00000163788
ENSDART00000171833 |
chd1
|
chromodomain helicase DNA binding protein 1 |
| chr2_+_52225622 | 0.36 |
ENSDART00000170353
|
ENSDARG00000103108
|
ENSDARG00000103108 |
| chr10_+_39971944 | 0.36 |
ENSDART00000003435
|
smfn
|
small fragment nuclease |
| chr23_+_25047201 | 0.36 |
ENSDART00000047020
|
casp9
|
caspase 9, apoptosis-related cysteine peptidase |
| chr17_+_11921705 | 0.36 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr11_-_30388949 | 0.36 |
ENSDART00000063775
|
ENSDARG00000043442
|
ENSDARG00000043442 |
| chr13_+_11306913 | 0.36 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr5_-_53776769 | 0.36 |
ENSDART00000164595
|
coq4
|
coenzyme Q4 homolog (S. cerevisiae) |
| chr14_-_46477165 | 0.36 |
ENSDART00000173209
ENSDART00000164837 |
ehd1a
|
EH-domain containing 1a |
| chr3_+_34876160 | 0.35 |
ENSDART00000029451
|
prkcbb
|
protein kinase C, beta b |
| chr20_+_19045423 | 0.34 |
ENSDART00000035447
|
mtmr9
|
myotubularin related protein 9 |
| chr24_+_12693997 | 0.34 |
|
|
|
| chr2_-_17820860 | 0.34 |
ENSDART00000024302
|
kdm4ab
|
lysine (K)-specific demethylase 4A, genome duplicate b |
| chr21_-_16442202 | 0.34 |
|
|
|
| chr10_-_6587375 | 0.34 |
ENSDART00000163788
ENSDART00000171833 |
chd1
|
chromodomain helicase DNA binding protein 1 |
| chr24_-_24884729 | 0.34 |
ENSDART00000168254
|
nup58
|
nucleoporin 58 |
| chr8_-_53593850 | 0.33 |
|
|
|
| chr4_-_12782246 | 0.33 |
ENSDART00000058020
|
helb
|
helicase (DNA) B |
| chr8_-_2471264 | 0.33 |
ENSDART00000056767
|
acaa2
|
acetyl-CoA acyltransferase 2 |
| chr13_+_11306998 | 0.33 |
ENSDART00000138312
|
zbtb18
|
zinc finger and BTB domain containing 18 |
| chr11_+_25302072 | 0.33 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr21_-_32748287 | 0.32 |
ENSDART00000031028
|
cnot6a
|
CCR4-NOT transcription complex, subunit 6a |
| chr10_+_36718872 | 0.32 |
ENSDART00000063359
|
ucp3
|
uncoupling protein 3 |
| chr19_-_17306333 | 0.31 |
ENSDART00000132662
|
stmn1a
|
stathmin 1a |
| chr7_+_23224924 | 0.31 |
ENSDART00000142401
|
zgc:109889
|
zgc:109889 |
| chr19_-_2314696 | 0.30 |
ENSDART00000166669
|
btd
|
biotinidase |
| chr23_+_16848213 | 0.30 |
ENSDART00000104791
|
zgc:153722
|
zgc:153722 |
| chr22_-_9790451 | 0.30 |
|
|
|
| chr5_-_22364806 | 0.30 |
ENSDART00000057541
|
apool
|
apolipoprotein O-like |
| chr19_+_33360835 | 0.30 |
ENSDART00000052091
|
atp6v1c1b
|
ATPase, H+ transporting, lysosomal, V1 subunit C1b |
| chr13_-_9034919 | 0.30 |
ENSDART00000135373
|
si:dkey-33c12.4
|
si:dkey-33c12.4 |
| chr22_+_1965847 | 0.30 |
|
|
|
| chr24_-_16835253 | 0.30 |
ENSDART00000005331
ENSDART00000128446 |
klhl15
|
kelch-like family member 15 |
| chr18_+_7584201 | 0.30 |
ENSDART00000163709
|
zgc:77650
|
zgc:77650 |
| chr11_+_11050173 | 0.29 |
|
|
|
| chr4_+_18427325 | 0.29 |
|
|
|
| chr16_+_38251653 | 0.29 |
ENSDART00000044971
|
myo1eb
|
myosin IE, b |
| chr3_+_34690515 | 0.29 |
|
|
|
| chr18_-_44366076 | 0.29 |
ENSDART00000166935
|
prdm10
|
PR domain containing 10 |
| chr18_+_20571785 | 0.29 |
ENSDART00000040074
|
wee2
|
WEE1 homolog 2 (S. pombe) |
| chr21_-_32747544 | 0.28 |
|
|
|
| chr19_+_42863138 | 0.28 |
ENSDART00000076938
|
pogza
|
pogo transposable element derived with ZNF domain a |
| chr1_+_52686829 | 0.28 |
ENSDART00000143909
|
tbc1d9
|
TBC1 domain family, member 9 (with GRAM domain) |
| chr24_+_26918580 | 0.28 |
ENSDART00000161327
|
larp4b
|
La ribonucleoprotein domain family, member 4B |
| chr19_-_35380449 | 0.28 |
|
|
|
| chr2_-_21954556 | 0.27 |
ENSDART00000145040
|
ralab
|
v-ral simian leukemia viral oncogene homolog Ab (ras related) |
| chr3_-_39048361 | 0.27 |
ENSDART00000013167
|
retsat
|
retinol saturase (all-trans-retinol 13,14-reductase) |
| chr14_+_11836698 | 0.27 |
ENSDART00000012438
|
hspa9
|
heat shock protein 9 |
| chr19_-_44469947 | 0.27 |
ENSDART00000006338
|
mterf3
|
mitochondrial transcription termination factor 3 |
| chr6_-_37534996 | 0.27 |
ENSDART00000147826
|
jade3
|
jade family PHD finger 3 |
| KN150699v1_-_7850 | 0.26 |
|
|
|
| chr17_+_12878081 | 0.26 |
ENSDART00000115208
|
fbxo33
|
F-box protein 33 |
| chr2_+_22753590 | 0.26 |
ENSDART00000171853
|
LMO4
|
zgc:56628 |
| chr2_-_26949759 | 0.26 |
|
|
|
| chr18_-_44366105 | 0.26 |
ENSDART00000166935
|
prdm10
|
PR domain containing 10 |
| chr10_+_187740 | 0.26 |
ENSDART00000167367
|
ets2
|
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
| chr7_+_23224800 | 0.25 |
ENSDART00000115299
ENSDART00000101423 |
zgc:109889
|
zgc:109889 |
| chr19_-_42988039 | 0.25 |
ENSDART00000128278
|
si:dkey-267n13.1
|
si:dkey-267n13.1 |
| chr22_+_2386743 | 0.25 |
ENSDART00000132925
|
zgc:112977
|
zgc:112977 |
| chr23_-_15280634 | 0.25 |
ENSDART00000056570
|
phf20b
|
PHD finger protein 20, b |
| chr17_+_18012036 | 0.24 |
ENSDART00000022758
|
setd3
|
SET domain containing 3 |
| chr23_-_27515964 | 0.24 |
ENSDART00000167838
|
dip2ba
|
disco-interacting protein 2 homolog Ba |
| chr16_+_38251717 | 0.23 |
ENSDART00000044971
|
myo1eb
|
myosin IE, b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.7 | GO:0060631 | regulation of meiosis I(GO:0060631) |
| 0.5 | 1.6 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.2 | 1.9 | GO:0035283 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.2 | 0.6 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) vascular smooth muscle contraction(GO:0014829) positive regulation of muscle contraction(GO:0045933) |
| 0.2 | 1.0 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 0.7 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.1 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 1.1 | GO:0060251 | regulation of glial cell proliferation(GO:0060251) |
| 0.1 | 0.9 | GO:0043536 | positive regulation of blood vessel endothelial cell migration(GO:0043536) positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.1 | 0.4 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.1 | 1.2 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 1.1 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.1 | 0.5 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.1 | 0.7 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 1.2 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.5 | GO:0021588 | cerebellum formation(GO:0021588) |
| 0.1 | 0.4 | GO:0090004 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.1 | 0.3 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.1 | 0.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.3 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.2 | GO:0070476 | rRNA (guanine-N7)-methylation(GO:0070476) |
| 0.1 | 0.2 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.1 | 0.4 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 0.6 | GO:0097324 | beta-amyloid metabolic process(GO:0050435) melanocyte migration(GO:0097324) |
| 0.1 | 0.4 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 0.2 | GO:0072003 | kidney rudiment formation(GO:0072003) pronephros formation(GO:0072116) |
| 0.1 | 1.1 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.1 | 0.3 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.6 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 1.5 | GO:0097205 | renal filtration(GO:0097205) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.7 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.0 | 0.7 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 0.2 | GO:0042373 | vitamin K metabolic process(GO:0042373) cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.4 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 0.6 | GO:0000959 | mitochondrial RNA metabolic process(GO:0000959) |
| 0.0 | 1.5 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.5 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 2.8 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.6 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.7 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.4 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.5 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0046755 | virion assembly(GO:0019068) virus maturation(GO:0019075) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.0 | 0.9 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.3 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.3 | GO:0042026 | protein refolding(GO:0042026) |
| 0.0 | 0.2 | GO:1901985 | positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.4 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.3 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:0010525 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.0 | 0.1 | GO:0097354 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.7 | GO:0032465 | regulation of cytokinesis(GO:0032465) |
| 0.0 | 0.9 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.0 | 0.5 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.2 | GO:0036302 | atrioventricular canal development(GO:0036302) |
| 0.0 | 0.4 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) quinone metabolic process(GO:1901661) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.3 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.4 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 1.6 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 0.3 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.3 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.3 | GO:0030537 | larval locomotory behavior(GO:0008345) larval behavior(GO:0030537) |
| 0.0 | 0.4 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.5 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.4 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.2 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.9 | GO:0051169 | nucleocytoplasmic transport(GO:0006913) nuclear transport(GO:0051169) |
| 0.0 | 0.5 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.1 | GO:0045003 | double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.2 | 1.1 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.2 | 1.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 0.4 | GO:0070390 | transcription export complex 2(GO:0070390) |
| 0.1 | 0.5 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 0.2 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 1.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.7 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.5 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 1.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.4 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 2.1 | GO:0031228 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 0.1 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.5 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.7 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 0.0 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.2 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 1.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.9 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0030175 | filopodium(GO:0030175) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.9 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.3 | 1.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.3 | 1.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.3 | 1.0 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.2 | 1.0 | GO:0070259 | tyrosyl-DNA phosphodiesterase activity(GO:0070259) |
| 0.1 | 0.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.1 | 0.6 | GO:0099528 | G-protein coupled neurotransmitter receptor activity(GO:0099528) |
| 0.1 | 0.4 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) |
| 0.1 | 1.7 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.4 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.1 | 1.6 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.1 | 0.3 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.1 | 0.4 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.1 | 0.5 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.1 | 3.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.5 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) efflux transmembrane transporter activity(GO:0015562) |
| 0.1 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.5 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 0.5 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.4 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.1 | 0.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 1.2 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.2 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.4 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.9 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.4 | GO:0022841 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.6 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.4 | GO:0097200 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.5 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.5 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 1.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.3 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.6 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.3 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.2 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.5 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.4 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.1 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 1.4 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 1.0 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.0 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.3 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.4 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 3.1 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.7 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.4 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.7 | GO:0003724 | RNA helicase activity(GO:0003724) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.7 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.7 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.7 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.0 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.9 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 1.0 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.5 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |