Project
DANIO-CODE
Navigation
Downloads
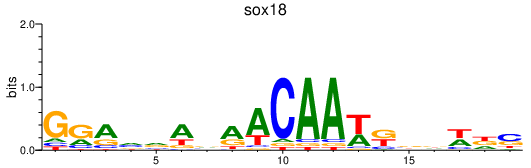
Results for sox18
Z-value: 0.73
Transcription factors associated with sox18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox18
|
ENSDARG00000058598 | SRY-box transcription factor 18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox18 | dr10_dc_chr23_+_8862155_8862175 | -0.66 | 5.3e-03 | Click! |
Activity profile of sox18 motif
Sorted Z-values of sox18 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of sox18
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_1522439 | 1.41 |
ENSDART00000175566
|
CABZ01085702.1
|
ENSDARG00000106927 |
| chr9_-_46614763 | 1.37 |
ENSDART00000009790
|
cx43.4
|
connexin 43.4 |
| chr19_+_15536640 | 1.20 |
ENSDART00000098970
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr14_-_23980261 | 0.89 |
ENSDART00000054243
|
dpf2l
|
D4, zinc and double PHD fingers family 2, like |
| chr4_+_13413303 | 0.89 |
ENSDART00000003694
|
cand1
|
cullin-associated and neddylation-dissociated 1 |
| chr14_+_29932533 | 0.88 |
ENSDART00000017122
|
asah1a
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1a |
| chr14_+_6122803 | 0.87 |
ENSDART00000149783
ENSDART00000148461 |
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr7_-_24724911 | 0.85 |
ENSDART00000173791
|
rcor2
|
REST corepressor 2 |
| chr16_-_39291647 | 0.85 |
ENSDART00000171342
|
tmem42a
|
transmembrane protein 42a |
| chr6_+_7916307 | 0.84 |
|
|
|
| chr4_-_13974345 | 0.83 |
ENSDART00000067177
ENSDART00000144127 |
prickle1b
|
prickle homolog 1b |
| chr14_-_23980060 | 0.81 |
ENSDART00000054243
|
dpf2l
|
D4, zinc and double PHD fingers family 2, like |
| chr20_+_34814623 | 0.77 |
ENSDART00000034252
|
pnocb
|
prepronociceptin b |
| chr17_+_25828037 | 0.77 |
|
|
|
| chr5_-_15971338 | 0.75 |
ENSDART00000110437
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr7_+_24543671 | 0.74 |
ENSDART00000173581
|
si:ch211-226l4.6
|
si:ch211-226l4.6 |
| chr16_+_31871645 | 0.73 |
ENSDART00000045210
|
mlf2
|
myeloid leukemia factor 2 |
| chr15_+_29795467 | 0.71 |
ENSDART00000099958
|
zgc:153372
|
zgc:153372 |
| chr11_+_17849608 | 0.71 |
ENSDART00000020283
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr6_+_49054117 | 0.71 |
ENSDART00000011876
|
sycp1
|
synaptonemal complex protein 1 |
| chr9_-_6861630 | 0.71 |
|
|
|
| chr21_-_34809442 | 0.70 |
ENSDART00000029708
|
zgc:56585
|
zgc:56585 |
| chr14_+_37205204 | 0.69 |
ENSDART00000173192
|
pcdh1b
|
protocadherin 1b |
| chr24_+_9335316 | 0.65 |
ENSDART00000132688
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr5_-_23697487 | 0.62 |
ENSDART00000013309
|
sox19a
|
SRY (sex determining region Y)-box 19a |
| chr1_+_45015350 | 0.60 |
ENSDART00000143363
|
si:ch211-214c7.4
|
si:ch211-214c7.4 |
| chr13_+_40689102 | 0.57 |
ENSDART00000016960
|
prkg1a
|
protein kinase, cGMP-dependent, type Ia |
| chr18_+_35253031 | 0.53 |
|
|
|
| chr25_+_34555948 | 0.53 |
ENSDART00000111706
|
si:dkey-108k21.14
|
si:dkey-108k21.14 |
| chr10_+_16071124 | 0.52 |
ENSDART00000065036
|
phax
|
phosphorylated adaptor for RNA export |
| chr9_+_45193290 | 0.52 |
ENSDART00000176175
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr25_-_34599241 | 0.52 |
ENSDART00000156727
|
si:dkey-108k21.21
|
si:dkey-108k21.21 |
| chr14_-_35060298 | 0.51 |
ENSDART00000129676
|
lsm11
|
LSM11, U7 small nuclear RNA associated |
| chr7_+_22582619 | 0.50 |
ENSDART00000148502
|
clcn5b
|
chloride channel, voltage-sensitive 5b |
| chr14_+_6655541 | 0.49 |
ENSDART00000148447
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr8_+_23705243 | 0.48 |
ENSDART00000142395
|
mkrn4
|
makorin, ring finger protein, 4 |
| chr7_+_22582561 | 0.47 |
ENSDART00000148502
|
clcn5b
|
chloride channel, voltage-sensitive 5b |
| chr19_+_10742387 | 0.45 |
ENSDART00000091813
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr14_+_6122901 | 0.44 |
ENSDART00000149783
ENSDART00000148461 |
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr2_-_48069574 | 0.44 |
ENSDART00000056306
|
ftr24
|
finTRIM family, member 24 |
| chr3_+_25868700 | 0.44 |
ENSDART00000024316
|
mcm5
|
minichromosome maintenance complex component 5 |
| chr14_+_8437594 | 0.42 |
|
|
|
| chr3_-_25246452 | 0.41 |
ENSDART00000177576
|
kpna2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
| chr22_+_24532560 | 0.38 |
ENSDART00000169847
|
wdr47b
|
WD repeat domain 47b |
| chr14_+_6656015 | 0.38 |
ENSDART00000148394
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr2_+_31959127 | 0.37 |
ENSDART00000132113
|
otulinb
|
OTU deubiquitinase with linear linkage specificity b |
| chr12_-_33256934 | 0.37 |
ENSDART00000066233
ENSDART00000148165 |
slc16a3
|
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr21_+_10152870 | 0.37 |
|
|
|
| chr21_-_4901827 | 0.36 |
ENSDART00000067733
|
zgc:77838
|
zgc:77838 |
| chr23_-_33692244 | 0.36 |
|
|
|
| chr4_+_46917725 | 0.36 |
|
|
|
| chr25_+_34556009 | 0.36 |
ENSDART00000111706
|
si:dkey-108k21.14
|
si:dkey-108k21.14 |
| chr23_-_10810190 | 0.35 |
ENSDART00000140745
|
eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr22_+_24532479 | 0.35 |
ENSDART00000169847
|
wdr47b
|
WD repeat domain 47b |
| chr14_+_6122644 | 0.34 |
ENSDART00000149783
ENSDART00000148461 |
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr25_-_22821132 | 0.34 |
|
|
|
| chr2_-_32279019 | 0.34 |
ENSDART00000159843
ENSDART00000056621 ENSDART00000039717 ENSDART00000145704 |
fam49ba
|
family with sequence similarity 49, member Ba |
| chr16_-_42015364 | 0.33 |
ENSDART00000136742
|
BX649388.1
|
ENSDARG00000091933 |
| chr24_-_31257458 | 0.32 |
ENSDART00000158808
|
cnn3a
|
calponin 3, acidic a |
| chr2_-_10173776 | 0.32 |
ENSDART00000137924
|
zgc:55943
|
zgc:55943 |
| chr7_-_24724452 | 0.31 |
ENSDART00000173791
|
rcor2
|
REST corepressor 2 |
| chr11_+_17849528 | 0.31 |
ENSDART00000020283
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr4_+_13413119 | 0.29 |
ENSDART00000003694
|
cand1
|
cullin-associated and neddylation-dissociated 1 |
| chr21_+_28710269 | 0.29 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr10_-_8083196 | 0.28 |
ENSDART00000059017
|
nfkbil1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
| chr1_+_48790583 | 0.28 |
ENSDART00000124833
|
pdcd11
|
programmed cell death 11 |
| chr8_-_24772008 | 0.27 |
ENSDART00000111617
|
rbm15
|
RNA binding motif protein 15 |
| chr3_-_25246374 | 0.27 |
ENSDART00000077538
|
kpna2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
| chr19_+_10742344 | 0.27 |
ENSDART00000091813
|
ago3b
|
argonaute RISC catalytic component 3b |
| chr5_-_67580755 | 0.26 |
ENSDART00000169350
|
ENSDARG00000098655
|
ENSDARG00000098655 |
| chr8_+_53202474 | 0.25 |
|
|
|
| chr4_-_70349899 | 0.25 |
|
|
|
| chr17_+_33862300 | 0.24 |
|
|
|
| chr8_+_17148864 | 0.24 |
ENSDART00000140531
|
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr3_+_25868480 | 0.24 |
ENSDART00000024316
|
mcm5
|
minichromosome maintenance complex component 5 |
| chr11_-_28367400 | 0.23 |
ENSDART00000065853
|
dhrs3b
|
dehydrogenase/reductase (SDR family) member 3b |
| chr24_+_9335428 | 0.23 |
ENSDART00000132688
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr21_-_43176006 | 0.23 |
ENSDART00000148630
|
hspa4a
|
heat shock protein 4a |
| chr24_-_31257591 | 0.23 |
ENSDART00000158808
|
cnn3a
|
calponin 3, acidic a |
| chr10_+_11439845 | 0.22 |
ENSDART00000064210
|
cwc27
|
CWC27 spliceosome-associated protein homolog (S. cerevisiae) |
| chr18_-_21759421 | 0.22 |
ENSDART00000079253
|
pskh1
|
protein serine kinase H1 |
| chr16_+_22949379 | 0.22 |
ENSDART00000137783
|
flad1
|
flavin adenine dinucleotide synthetase 1 |
| chr14_+_6655894 | 0.21 |
ENSDART00000148394
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr10_+_22921629 | 0.21 |
ENSDART00000057291
|
arrb2a
|
arrestin, beta 2a |
| chr12_+_19078444 | 0.21 |
|
|
|
| chr15_+_29795509 | 0.21 |
ENSDART00000099958
|
zgc:153372
|
zgc:153372 |
| chr17_+_51593393 | 0.19 |
ENSDART00000004379
|
nol10
|
nucleolar protein 10 |
| chr12_-_35004566 | 0.17 |
ENSDART00000153034
|
si:ch73-127m5.2
|
si:ch73-127m5.2 |
| chr4_-_29167253 | 0.17 |
ENSDART00000160579
|
CR388132.2
|
ENSDARG00000101782 |
| chr9_+_32496059 | 0.17 |
ENSDART00000078513
|
mob4
|
MOB family member 4, phocein |
| chr16_+_22949494 | 0.16 |
ENSDART00000137783
|
flad1
|
flavin adenine dinucleotide synthetase 1 |
| chr8_+_53202445 | 0.15 |
|
|
|
| chr3_-_23340407 | 0.15 |
ENSDART00000113135
ENSDART00000166068 |
msl1a
|
male-specific lethal 1 homolog a (Drosophila) |
| chr2_+_31959275 | 0.15 |
ENSDART00000132113
|
otulinb
|
OTU deubiquitinase with linear linkage specificity b |
| chr8_+_14120357 | 0.14 |
ENSDART00000080832
|
si:dkey-6n6.2
|
si:dkey-6n6.2 |
| chr9_+_56880744 | 0.14 |
|
|
|
| chr24_+_20228016 | 0.14 |
ENSDART00000139989
|
plcd1a
|
phospholipase C, delta 1a |
| chr9_-_33975936 | 0.12 |
ENSDART00000140779
|
fundc1
|
FUN14 domain containing 1 |
| chr17_+_50484982 | 0.11 |
ENSDART00000075150
|
bmp4
|
bone morphogenetic protein 4 |
| chr11_-_43409685 | 0.11 |
ENSDART00000163843
|
zgc:153431
|
zgc:153431 |
| chr2_+_31959316 | 0.10 |
ENSDART00000132113
|
otulinb
|
OTU deubiquitinase with linear linkage specificity b |
| chr9_+_37945009 | 0.09 |
ENSDART00000100592
|
pdia5
|
protein disulfide isomerase family A, member 5 |
| chr5_+_39066924 | 0.09 |
ENSDART00000085388
|
bmp3
|
bone morphogenetic protein 3 |
| chr17_-_14772485 | 0.08 |
ENSDART00000080401
ENSDART00000154690 |
si:ch211-266o15.1
|
si:ch211-266o15.1 |
| chr2_-_28244864 | 0.08 |
ENSDART00000137542
|
zgc:163121
|
zgc:163121 |
| chr11_-_27260783 | 0.08 |
|
|
|
| chr24_-_40968527 | 0.07 |
ENSDART00000172617
|
smyhc1
|
slow myosin heavy chain 1 |
| chr17_+_48929046 | 0.07 |
ENSDART00000171615
|
CU499333.1
|
ENSDARG00000103628 |
| chr1_-_49967146 | 0.07 |
ENSDART00000015591
|
ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr9_-_43015354 | 0.06 |
|
|
|
| chr14_-_35060332 | 0.06 |
ENSDART00000129676
|
lsm11
|
LSM11, U7 small nuclear RNA associated |
| chr5_-_12706568 | 0.05 |
ENSDART00000051666
|
ppm1f
|
protein phosphatase, Mg2+/Mn2+ dependent, 1F |
| chr9_+_45031009 | 0.05 |
ENSDART00000002633
|
frzb
|
frizzled-related protein |
| chr23_-_10810264 | 0.04 |
ENSDART00000013768
|
eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr8_+_23704956 | 0.04 |
ENSDART00000132734
|
mkrn4
|
makorin, ring finger protein, 4 |
| chr5_-_23013841 | 0.04 |
ENSDART00000137655
|
stag2a
|
stromal antigen 2a |
| KN150118v1_-_40245 | 0.04 |
|
|
|
| chr5_-_30104184 | 0.03 |
ENSDART00000016709
|
ift22
|
intraflagellar transport 22 homolog (Chlamydomonas) |
| chr13_-_44926708 | 0.03 |
ENSDART00000130467
ENSDART00000074791 ENSDART00000136679 |
chp1
|
calcineurin-like EF-hand protein 1 |
| chr2_+_21527785 | 0.02 |
ENSDART00000136498
|
si:dkey-29d8.3
|
si:dkey-29d8.3 |
| chr24_-_24578984 | 0.02 |
ENSDART00000012399
|
armc1
|
armadillo repeat containing 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.2 | 0.7 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.2 | 0.7 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.2 | 0.9 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.2 | 0.5 | GO:0006408 | snRNA export from nucleus(GO:0006408) |
| 0.1 | 1.2 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.1 | 1.0 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.1 | 0.4 | GO:0046443 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.1 | 0.8 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 0.6 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.6 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.2 | GO:0043393 | G-protein coupled receptor internalization(GO:0002031) regulation of protein binding(GO:0043393) |
| 0.1 | 0.7 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.1 | 0.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 1.4 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.7 | GO:0072091 | regulation of stem cell proliferation(GO:0072091) |
| 0.0 | 0.8 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.1 | GO:0035122 | embryonic medial fin morphogenesis(GO:0035122) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 1.2 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 1.0 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.3 | GO:0034112 | positive regulation of homotypic cell-cell adhesion(GO:0034112) positive regulation of T cell activation(GO:0050870) positive regulation of leukocyte cell-cell adhesion(GO:1903039) |
| 0.0 | 0.7 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.6 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.7 | GO:0071546 | pi-body(GO:0071546) |
| 0.1 | 0.6 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.4 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 1.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 0.7 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.8 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.7 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 1.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 1.4 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.5 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0030791 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.2 | 0.9 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.2 | 1.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.2 | 0.6 | GO:0030553 | cGMP-dependent protein kinase activity(GO:0004692) cGMP binding(GO:0030553) |
| 0.2 | 0.7 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.7 | GO:0090624 | endoribonuclease activity, cleaving miRNA-paired mRNA(GO:0090624) |
| 0.1 | 0.4 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.1 | 1.0 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 1.0 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.8 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.1 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.7 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.7 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.7 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 1.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.4 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.6 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |