Project
DANIO-CODE
Navigation
Downloads
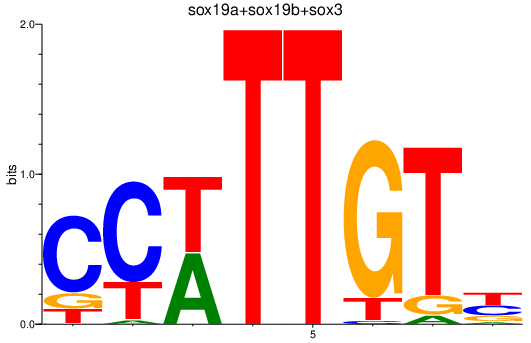
Results for sox19a+sox19b+sox3
Z-value: 2.92
Transcription factors associated with sox19a+sox19b+sox3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox19a
|
ENSDARG00000010770 | SRY-box transcription factor 19a |
|
sox19b
|
ENSDARG00000040266 | SRY-box transcription factor 19b |
|
sox3
|
ENSDARG00000053569 | SRY-box transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox19b | dr10_dc_chr7_-_26226451_26226578 | -0.97 | 1.2e-09 | Click! |
| sox19a | dr10_dc_chr5_-_23697487_23697817 | 0.80 | 2.3e-04 | Click! |
| sox3 | dr10_dc_chr14_-_32404076_32404177 | 0.75 | 7.5e-04 | Click! |
Activity profile of sox19a+sox19b+sox3 motif
Sorted Z-values of sox19a+sox19b+sox3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of sox19a+sox19b+sox3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_12733209 | 8.94 |
ENSDART00000167021
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr10_+_33227967 | 8.37 |
ENSDART00000159666
|
myl10
|
myosin, light chain 10, regulatory |
| chr9_-_42894582 | 8.35 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr1_+_16683931 | 8.14 |
ENSDART00000103262
ENSDART00000145068 ENSDART00000169619 ENSDART00000010526 |
fat1a
|
FAT atypical cadherin 1a |
| chr10_+_18994733 | 7.70 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr19_-_18316262 | 7.50 |
ENSDART00000151133
|
top2b
|
topoisomerase (DNA) II beta |
| chr12_-_30662709 | 7.24 |
ENSDART00000126466
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr2_-_43110857 | 7.20 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr13_-_13163676 | 6.75 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr1_+_44205158 | 6.49 |
ENSDART00000142820
|
wu:fc21g02
|
wu:fc21g02 |
| chr16_+_23482744 | 6.43 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr1_-_48853800 | 6.43 |
ENSDART00000137357
|
zgc:175214
|
zgc:175214 |
| chr5_+_34857986 | 6.42 |
ENSDART00000141239
|
erlin2
|
ER lipid raft associated 2 |
| chr20_+_31366832 | 6.33 |
ENSDART00000133353
|
apobb.1
|
apolipoprotein Bb, tandem duplicate 1 |
| chr6_-_43094573 | 6.24 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr23_+_21610798 | 6.09 |
ENSDART00000111966
|
arhgef10lb
|
Rho guanine nucleotide exchange factor (GEF) 10-like b |
| chr14_-_36037883 | 6.01 |
ENSDART00000173006
|
gpm6aa
|
glycoprotein M6Aa |
| chr8_-_16662185 | 5.97 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr19_+_5399813 | 5.75 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr18_+_22617072 | 5.49 |
ENSDART00000128965
|
bcar1
|
breast cancer anti-estrogen resistance 1 |
| chr23_+_32573474 | 5.44 |
ENSDART00000134811
|
si:dkey-261h17.1
|
si:dkey-261h17.1 |
| chr10_+_21850059 | 5.43 |
ENSDART00000164634
ENSDART00000172513 |
pcdh1g32
|
protocadherin 1 gamma 32 |
| chr21_+_27346176 | 5.37 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr21_-_35806638 | 5.33 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr19_-_19888074 | 5.31 |
|
|
|
| chr14_+_11151485 | 5.27 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr16_+_24045774 | 5.19 |
ENSDART00000133484
|
apoeb
|
apolipoprotein Eb |
| chr24_+_10276926 | 5.18 |
ENSDART00000145771
ENSDART00000157350 |
CT573344.1
|
ENSDARG00000093108 |
| chr7_-_57030751 | 5.06 |
|
|
|
| chr23_+_36832325 | 4.95 |
|
|
|
| chr13_+_22545318 | 4.95 |
ENSDART00000143312
|
zgc:193505
|
zgc:193505 |
| chr14_-_7774262 | 4.84 |
ENSDART00000045109
|
zgc:92242
|
zgc:92242 |
| chr11_-_41357639 | 4.81 |
ENSDART00000055709
|
her2
|
hairy-related 2 |
| chr20_-_20412830 | 4.74 |
ENSDART00000114779
|
ENSDARG00000079369
|
ENSDARG00000079369 |
| chr11_-_43331509 | 4.61 |
|
|
|
| chr3_+_30790513 | 4.60 |
ENSDART00000130422
|
cldni
|
claudin i |
| chr4_-_8610868 | 4.59 |
ENSDART00000067322
|
fbxl14b
|
F-box and leucine-rich repeat protein 14b |
| chr24_-_33869817 | 4.57 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr5_-_36349284 | 4.55 |
ENSDART00000047269
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr24_-_40968409 | 4.54 |
ENSDART00000169315
ENSDART00000171543 |
smyhc1
|
slow myosin heavy chain 1 |
| chr13_+_28601627 | 4.52 |
ENSDART00000015773
|
ldb1a
|
LIM domain binding 1a |
| chr23_+_35609887 | 4.52 |
ENSDART00000179393
|
tuba1b
|
tubulin, alpha 1b |
| chr5_-_33006537 | 4.48 |
ENSDART00000048350
|
si:dkey-34e4.1
|
si:dkey-34e4.1 |
| chr24_-_41448993 | 4.47 |
ENSDART00000041349
|
crygn2
|
crystallin, gamma N2 |
| chr23_+_23730717 | 4.45 |
|
|
|
| chr2_-_59531831 | 4.42 |
ENSDART00000168568
|
CU861477.1
|
ENSDARG00000100128 |
| chr16_+_34569479 | 4.41 |
ENSDART00000041007
|
stmn1b
|
stathmin 1b |
| chr1_+_46537108 | 4.40 |
|
|
|
| chr13_+_25298383 | 4.40 |
|
|
|
| chr23_-_7892862 | 4.34 |
ENSDART00000157612
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr13_-_13163801 | 4.33 |
ENSDART00000156968
|
fgfr3
|
fibroblast growth factor receptor 3 |
| chr7_-_7442720 | 4.31 |
ENSDART00000091105
|
mfap3l
|
microfibrillar-associated protein 3-like |
| chr6_+_47845002 | 4.25 |
ENSDART00000140943
|
padi2
|
peptidyl arginine deiminase, type II |
| chr5_-_14148028 | 4.23 |
ENSDART00000113037
|
sema4c
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr19_-_18316039 | 4.23 |
ENSDART00000151133
|
top2b
|
topoisomerase (DNA) II beta |
| chr23_-_30119058 | 4.19 |
ENSDART00000103480
|
ccdc187
|
coiled-coil domain containing 187 |
| chr11_+_24479085 | 4.19 |
ENSDART00000145217
|
ENSDARG00000070571
|
ENSDARG00000070571 |
| chr25_+_17774363 | 4.11 |
ENSDART00000136797
|
btbd10a
|
BTB (POZ) domain containing 10a |
| chr14_-_26406720 | 4.11 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr3_-_16100715 | 4.10 |
ENSDART00000146699
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr2_+_20118917 | 4.08 |
ENSDART00000038648
|
ptbp2b
|
polypyrimidine tract binding protein 2b |
| chr22_+_17091411 | 4.07 |
ENSDART00000170076
|
frem1b
|
Fras1 related extracellular matrix 1b |
| chr8_-_12171624 | 3.96 |
ENSDART00000132824
|
dab2ipa
|
DAB2 interacting protein a |
| chr19_+_12995955 | 3.95 |
ENSDART00000132892
|
cthrc1a
|
collagen triple helix repeat containing 1a |
| chr7_+_39173765 | 3.94 |
ENSDART00000173748
|
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr7_-_7164950 | 3.93 |
ENSDART00000012637
|
zgc:101810
|
zgc:101810 |
| chr23_-_7889744 | 3.92 |
ENSDART00000164117
|
myt1b
|
myelin transcription factor 1b |
| chr2_-_30675594 | 3.92 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr22_-_37417903 | 3.91 |
ENSDART00000149948
|
FP102167.1
|
ENSDARG00000095844 |
| chr14_-_40918015 | 3.87 |
ENSDART00000147389
|
tmem35
|
transmembrane protein 35 |
| chr1_-_50147413 | 3.87 |
ENSDART00000080389
|
fam13a
|
family with sequence similarity 13, member A |
| chr6_+_29800606 | 3.87 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr17_-_36988937 | 3.82 |
ENSDART00000145236
|
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr7_-_32888309 | 3.81 |
ENSDART00000173461
|
BX571811.2
|
ENSDARG00000105655 |
| chr18_-_15498798 | 3.79 |
ENSDART00000109410
|
endouc
|
endonuclease, polyU-specific C |
| chr20_+_34867305 | 3.70 |
ENSDART00000018304
|
mcm3
|
minichromosome maintenance complex component 3 |
| chr23_+_20183765 | 3.70 |
ENSDART00000054664
|
tnnc1b
|
troponin C type 1b (slow) |
| chr25_+_19992389 | 3.61 |
ENSDART00000143441
|
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| KN150173v1_-_16439 | 3.60 |
ENSDART00000162810
|
SNORD60
|
Small nucleolar RNA SNORD60 |
| chr6_+_27156169 | 3.59 |
ENSDART00000088364
|
kif1aa
|
kinesin family member 1Aa |
| chr12_-_36138709 | 3.58 |
ENSDART00000130985
|
st6galnac
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase |
| chr11_+_37345024 | 3.57 |
ENSDART00000077496
|
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr25_-_22089794 | 3.56 |
ENSDART00000139110
|
pkp3a
|
plakophilin 3a |
| chr1_-_34921848 | 3.56 |
ENSDART00000142154
|
frem3
|
Fras1 related extracellular matrix 3 |
| chr18_+_40535215 | 3.55 |
ENSDART00000132833
|
vwa7
|
von Willebrand factor A domain containing 7 |
| chr6_-_12616971 | 3.54 |
ENSDART00000056764
|
bmpr2a
|
bone morphogenetic protein receptor, type II a (serine/threonine kinase) |
| chr5_-_54890685 | 3.53 |
ENSDART00000148436
|
gna14
|
guanine nucleotide binding protein (G protein), alpha 14 |
| chr21_+_25651712 | 3.52 |
ENSDART00000135123
|
si:dkey-17e16.17
|
si:dkey-17e16.17 |
| chr6_-_43094926 | 3.52 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr13_+_27102308 | 3.52 |
ENSDART00000145901
|
rin2
|
Ras and Rab interactor 2 |
| chr11_+_30048913 | 3.49 |
|
|
|
| chr6_+_54231519 | 3.43 |
ENSDART00000149542
|
pacsin1b
|
protein kinase C and casein kinase substrate in neurons 1b |
| chr15_-_4537178 | 3.42 |
ENSDART00000155619
ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| KN150230v1_-_36595 | 3.37 |
|
|
|
| chr5_-_45905518 | 3.37 |
ENSDART00000111589
|
hapln1a
|
hyaluronan and proteoglycan link protein 1a |
| chr9_-_33584190 | 3.36 |
ENSDART00000138844
|
caska
|
calcium/calmodulin-dependent serine protein kinase a |
| chr13_+_27102377 | 3.36 |
ENSDART00000138043
|
rin2
|
Ras and Rab interactor 2 |
| chr10_+_44195410 | 3.35 |
ENSDART00000046172
|
cryba4
|
crystallin, beta A4 |
| chr2_+_3125365 | 3.35 |
ENSDART00000164308
ENSDART00000167649 |
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr10_+_31362144 | 3.35 |
|
|
|
| chr25_+_30746445 | 3.34 |
ENSDART00000156916
|
lsp1
|
lymphocyte-specific protein 1 |
| chr10_-_36690119 | 3.32 |
ENSDART00000077161
|
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 |
| chr8_+_23071884 | 3.32 |
ENSDART00000063075
|
zgc:100920
|
zgc:100920 |
| chr6_+_30441419 | 3.31 |
|
|
|
| chr16_-_38117896 | 3.30 |
ENSDART00000114266
|
plekho1b
|
pleckstrin homology domain containing, family O member 1b |
| chr12_-_7669319 | 3.25 |
|
|
|
| chr16_-_32022177 | 3.25 |
ENSDART00000139664
|
styk1
|
serine/threonine/tyrosine kinase 1 |
| chr22_-_3264630 | 3.23 |
ENSDART00000170992
|
si:zfos-943e10.1
|
si:zfos-943e10.1 |
| chr9_+_4188263 | 3.23 |
ENSDART00000164543
|
kalrna
|
kalirin RhoGEF kinase a |
| chr11_+_24687813 | 3.17 |
ENSDART00000131431
|
sulf2a
|
sulfatase 2a |
| chr9_+_41301317 | 3.14 |
ENSDART00000000250
|
slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr13_+_11305781 | 3.13 |
|
|
|
| chr19_+_5399877 | 3.12 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr7_+_23843682 | 3.12 |
ENSDART00000173899
|
mrpl52
|
mitochondrial ribosomal protein L52 |
| chr12_+_3043444 | 3.11 |
ENSDART00000149427
|
sgca
|
sarcoglycan, alpha |
| chr5_+_15167637 | 3.11 |
ENSDART00000127015
|
srrm4
|
serine/arginine repetitive matrix 4 |
| chr6_+_30681170 | 3.09 |
ENSDART00000112294
|
ttc22
|
tetratricopeptide repeat domain 22 |
| chr14_+_23980348 | 3.07 |
ENSDART00000160984
|
BX005228.1
|
ENSDARG00000105465 |
| chr1_+_45063098 | 3.07 |
ENSDART00000084512
|
pkn1a
|
protein kinase N1a |
| chr5_+_64411412 | 3.07 |
ENSDART00000158484
|
BX001023.2
|
ENSDARG00000101163 |
| chr21_-_20674965 | 3.05 |
ENSDART00000065649
|
ENSDARG00000044676
|
ENSDARG00000044676 |
| chr3_-_12032297 | 3.00 |
ENSDART00000140123
|
hmox2b
|
heme oxygenase 2b |
| chr5_-_65349550 | 3.00 |
ENSDART00000164228
|
nrarpb
|
notch-regulated ankyrin repeat protein b |
| chr14_+_34146377 | 2.99 |
ENSDART00000131861
|
tmsb2
|
thymosin beta 2 |
| chr1_+_39844690 | 2.98 |
ENSDART00000122059
|
scoca
|
short coiled-coil protein a |
| chr4_+_5733160 | 2.97 |
ENSDART00000110243
|
pou3f2a
|
POU class 3 homeobox 2a |
| chr12_-_7572970 | 2.96 |
ENSDART00000158095
|
slc16a9b
|
solute carrier family 16, member 9b |
| chr13_-_869704 | 2.93 |
|
|
|
| chr17_+_16557246 | 2.92 |
ENSDART00000015729
ENSDART00000136874 |
foxn3
|
forkhead box N3 |
| chr3_+_28450576 | 2.89 |
ENSDART00000150893
|
sept12
|
septin 12 |
| chr14_+_47326080 | 2.88 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr15_-_31687 | 2.87 |
|
|
|
| chr5_-_31275147 | 2.85 |
ENSDART00000098160
|
tmem119b
|
transmembrane protein 119b |
| chr14_-_46407410 | 2.84 |
ENSDART00000173078
|
si:ch211-168f7.5
|
si:ch211-168f7.5 |
| chr8_+_17148864 | 2.83 |
ENSDART00000140531
|
dimt1l
|
DIM1 dimethyladenosine transferase 1-like (S. cerevisiae) |
| chr10_-_27261937 | 2.82 |
|
|
|
| chr3_+_17387551 | 2.81 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr13_+_1051015 | 2.80 |
ENSDART00000033528
|
tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr3_+_14238188 | 2.78 |
ENSDART00000165452
ENSDART00000171726 |
tmem56b
|
transmembrane protein 56b |
| chr6_-_13654186 | 2.78 |
ENSDART00000150102
ENSDART00000041269 |
cryba2a
|
crystallin, beta A2a |
| chr10_+_4717800 | 2.77 |
ENSDART00000161789
|
palm2
|
paralemmin 2 |
| chr2_-_59047356 | 2.77 |
|
|
|
| chr10_+_22566353 | 2.75 |
ENSDART00000079514
|
efnb3a
|
ephrin-B3a |
| chr20_+_23599157 | 2.75 |
ENSDART00000149922
|
palld
|
palladin, cytoskeletal associated protein |
| chr4_+_4223606 | 2.74 |
|
|
|
| chr24_-_24306469 | 2.74 |
ENSDART00000154149
|
BX323067.1
|
ENSDARG00000097984 |
| chr19_+_15538967 | 2.73 |
ENSDART00000171403
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr7_-_26036887 | 2.71 |
ENSDART00000146935
|
zgc:77439
|
zgc:77439 |
| chr17_-_29102320 | 2.71 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr11_+_15478585 | 2.71 |
ENSDART00000163289
ENSDART00000066033 |
gdf11
|
growth differentiation factor 11 |
| chr14_+_38445969 | 2.71 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr24_+_20414578 | 2.70 |
ENSDART00000131677
|
hhatlb
|
hedgehog acyltransferase-like, b |
| chr11_+_41401893 | 2.70 |
ENSDART00000171655
|
AL929185.1
|
ENSDARG00000103982 |
| chr2_-_31037043 | 2.69 |
ENSDART00000087026
|
emilin2a
|
elastin microfibril interfacer 2a |
| chr4_+_18854068 | 2.67 |
ENSDART00000066977
|
bik
|
BCL2-interacting killer (apoptosis-inducing) |
| chr5_+_64411584 | 2.65 |
ENSDART00000158484
|
BX001023.2
|
ENSDARG00000101163 |
| chr17_+_25503946 | 2.63 |
|
|
|
| chr7_+_20272091 | 2.63 |
ENSDART00000052917
|
slc3a2a
|
solute carrier family 3 (amino acid transporter heavy chain), member 2a |
| chr15_-_16271208 | 2.62 |
|
|
|
| chr7_-_69399273 | 2.61 |
ENSDART00000159823
ENSDART00000075178 ENSDART00000168942 ENSDART00000126739 |
tspan5a
|
tetraspanin 5a |
| chr8_-_13503775 | 2.61 |
ENSDART00000063834
|
ENSDARG00000043482
|
ENSDARG00000043482 |
| chr23_-_43595189 | 2.61 |
|
|
|
| chr14_-_425659 | 2.60 |
|
|
|
| chr2_-_24614261 | 2.60 |
ENSDART00000099532
|
vmhc
|
ventricular myosin heavy chain |
| chr3_-_30027552 | 2.58 |
ENSDART00000103502
|
si:ch211-152f23.5
|
si:ch211-152f23.5 |
| chr5_-_51109419 | 2.57 |
ENSDART00000163464
|
lhfpl2b
|
lipoma HMGIC fusion partner-like 2b |
| chr20_+_20600211 | 2.57 |
ENSDART00000036124
|
six1b
|
SIX homeobox 1b |
| chr5_-_40310638 | 2.56 |
ENSDART00000133183
|
parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr16_+_51317933 | 2.53 |
ENSDART00000157736
|
hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr9_-_22311713 | 2.53 |
ENSDART00000108748
|
crygm2d18
|
crystallin, gamma M2d18 |
| chr23_-_18945009 | 2.49 |
ENSDART00000080064
|
CR847953.1
|
ENSDARG00000057403 |
| chr2_+_56339323 | 2.49 |
ENSDART00000123392
|
rab11bb
|
RAB11B, member RAS oncogene family, b |
| chr2_+_38178934 | 2.48 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr15_-_39756101 | 2.48 |
|
|
|
| chr3_+_19096030 | 2.47 |
ENSDART00000134514
|
smarca4a
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4a |
| chr18_+_44656323 | 2.46 |
ENSDART00000059063
|
ehd2b
|
EH-domain containing 2b |
| chr19_-_20148533 | 2.46 |
ENSDART00000163790
|
evx1
|
even-skipped homeobox 1 |
| chr9_-_13991784 | 2.45 |
ENSDART00000061156
|
raph1a
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1a |
| chr10_-_42111408 | 2.45 |
ENSDART00000033121
|
morn3
|
MORN repeat containing 3 |
| chr3_-_21217799 | 2.44 |
ENSDART00000114906
|
fam171a2a
|
family with sequence similarity 171, member A2a |
| chr9_-_22371512 | 2.44 |
ENSDART00000101809
|
crygm2d6
|
crystallin, gamma M2d6 |
| chr13_-_31310439 | 2.43 |
ENSDART00000076571
|
rtn1a
|
reticulon 1a |
| chr17_-_44133941 | 2.43 |
ENSDART00000126097
|
otx2
|
orthodenticle homeobox 2 |
| chr17_-_6219019 | 2.42 |
|
|
|
| chr8_-_38168395 | 2.42 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr24_+_24924379 | 2.42 |
ENSDART00000115165
|
amer2
|
APC membrane recruitment protein 2 |
| chr8_+_14120313 | 2.41 |
ENSDART00000080832
|
si:dkey-6n6.2
|
si:dkey-6n6.2 |
| chr9_-_3700395 | 2.41 |
ENSDART00000102900
|
sp5a
|
Sp5 transcription factor a |
| chr19_+_5375413 | 2.41 |
ENSDART00000141237
|
si:dkeyp-113d7.10
|
si:dkeyp-113d7.10 |
| chr20_+_26408687 | 2.40 |
|
|
|
| chr6_+_29420644 | 2.39 |
ENSDART00000065293
|
usp13
|
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chr3_-_40263048 | 2.38 |
ENSDART00000154236
|
AL928657.1
|
ENSDARG00000097119 |
| chr5_-_30015572 | 2.38 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr20_+_53310252 | 2.36 |
ENSDART00000175125
|
CABZ01089116.1
|
ENSDARG00000106770 |
| chr1_+_45503061 | 2.34 |
ENSDART00000010894
|
sox1b
|
SRY (sex determining region Y)-box 1b |
| chr17_-_35011682 | 2.33 |
|
|
|
| chr8_-_18502159 | 2.32 |
ENSDART00000148802
ENSDART00000149081 ENSDART00000148962 |
nexn
|
nexilin (F actin binding protein) |
| chr15_+_28435937 | 2.32 |
ENSDART00000142298
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr16_+_16941228 | 2.31 |
ENSDART00000142155
|
myh14
|
myosin, heavy chain 14, non-muscle |
| chr15_-_18110169 | 2.31 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 8.3 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 1.9 | 5.8 | GO:0071831 | chylomicron remnant clearance(GO:0034382) triglyceride-rich lipoprotein particle clearance(GO:0071830) intermediate-density lipoprotein particle clearance(GO:0071831) |
| 1.6 | 9.7 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 1.4 | 4.3 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 1.4 | 9.8 | GO:0021553 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 1.4 | 5.5 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 1.3 | 11.8 | GO:0007412 | axon target recognition(GO:0007412) |
| 1.2 | 7.2 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 1.2 | 13.2 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 1.1 | 12.5 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 1.1 | 5.5 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 1.1 | 12.6 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) mitotic DNA replication(GO:1902969) |
| 1.0 | 7.2 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 1.0 | 4.1 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.9 | 2.8 | GO:0001659 | temperature homeostasis(GO:0001659) sleep(GO:0030431) |
| 0.9 | 2.8 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.9 | 2.7 | GO:0050748 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.8 | 3.4 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.8 | 4.8 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.8 | 3.0 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.7 | 5.9 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.7 | 2.8 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.7 | 0.7 | GO:1903429 | regulation of cell maturation(GO:1903429) |
| 0.7 | 2.1 | GO:0097037 | heme export(GO:0097037) |
| 0.7 | 4.0 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.6 | 4.2 | GO:0060579 | ventral spinal cord interneuron fate commitment(GO:0060579) |
| 0.6 | 2.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.6 | 4.5 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.5 | 2.1 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.5 | 3.0 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.5 | 3.0 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.5 | 3.4 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.5 | 7.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.5 | 6.3 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.5 | 1.9 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.5 | 3.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.4 | 3.1 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.4 | 8.0 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.4 | 29.5 | GO:0007601 | visual perception(GO:0007601) |
| 0.4 | 4.7 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.4 | 0.8 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.4 | 2.3 | GO:0045056 | transcytosis(GO:0045056) |
| 0.4 | 7.6 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.4 | 0.7 | GO:0043282 | pharyngeal muscle development(GO:0043282) |
| 0.4 | 2.2 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.4 | 3.2 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.4 | 12.4 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.3 | 3.1 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.3 | 3.9 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.3 | 1.3 | GO:0010226 | response to lithium ion(GO:0010226) |
| 0.3 | 2.5 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.3 | 0.9 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.3 | 0.9 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.3 | 2.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.3 | 5.4 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.3 | 6.5 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.3 | 1.7 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.3 | 3.2 | GO:0043584 | nose development(GO:0043584) |
| 0.2 | 3.0 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.2 | 2.0 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.2 | 0.9 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.2 | 3.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.2 | 0.9 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.2 | 2.3 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.2 | 0.7 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) hypochord development(GO:0055016) radial glial cell differentiation(GO:0060019) |
| 0.2 | 3.4 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.2 | 3.9 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.2 | 1.7 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 0.6 | GO:0036058 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 2.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 2.0 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.2 | 0.6 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.2 | 1.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 0.7 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.2 | 1.8 | GO:0044030 | regulation of DNA methylation(GO:0044030) |
| 0.2 | 0.5 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.2 | 0.8 | GO:0006582 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.2 | 0.9 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 2.8 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.1 | 1.5 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 4.0 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.1 | 0.6 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.1 | 0.8 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 2.0 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.1 | 4.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 0.5 | GO:0043981 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.1 | 0.8 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.4 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.1 | 3.2 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.1 | 1.8 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 0.9 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 3.5 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 0.6 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.1 | 2.7 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 2.0 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 1.6 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.1 | 2.0 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.1 | 1.9 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 1.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 1.1 | GO:0042551 | neuron maturation(GO:0042551) |
| 0.1 | 0.6 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) regulation of mitotic attachment of spindle microtubules to kinetochore(GO:1902423) positive regulation of attachment of mitotic spindle microtubules to kinetochore(GO:1902425) RNA localization to chromatin(GO:1990280) |
| 0.1 | 4.0 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 1.9 | GO:0032944 | regulation of mononuclear cell proliferation(GO:0032944) regulation of T cell proliferation(GO:0042129) regulation of lymphocyte proliferation(GO:0050670) regulation of leukocyte proliferation(GO:0070663) |
| 0.1 | 1.7 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 1.3 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.1 | 2.1 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 3.2 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 0.8 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 0.8 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.1 | 3.2 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 4.6 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 0.7 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.1 | 3.1 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.1 | 2.0 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 1.0 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.8 | GO:0009649 | entrainment of circadian clock(GO:0009649) entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.1 | 2.4 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.1 | 1.3 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.1 | 2.2 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 1.9 | GO:0060401 | cytosolic calcium ion transport(GO:0060401) calcium ion transport into cytosol(GO:0060402) |
| 0.1 | 0.6 | GO:0003319 | cardioblast migration to the midline involved in heart rudiment formation(GO:0003319) |
| 0.1 | 3.2 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 2.4 | GO:0048332 | mesoderm morphogenesis(GO:0048332) |
| 0.1 | 1.5 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.8 | GO:0021681 | cerebellar granular layer development(GO:0021681) cerebellar granular layer morphogenesis(GO:0021683) cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 3.0 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.1 | 1.8 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 0.9 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.1 | 3.2 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.1 | 0.2 | GO:0061400 | positive regulation of transcription from RNA polymerase II promoter in response to calcium ion(GO:0061400) |
| 0.1 | 0.8 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.1 | 1.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 5.3 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.2 | GO:0045665 | negative regulation of neuron differentiation(GO:0045665) |
| 0.0 | 0.8 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 1.8 | GO:0043280 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043280) |
| 0.0 | 0.5 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.0 | 2.4 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.6 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.0 | 2.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 4.9 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 1.6 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 1.2 | GO:1901880 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) negative regulation of protein complex disassembly(GO:0043242) actin filament capping(GO:0051693) regulation of protein depolymerization(GO:1901879) negative regulation of protein depolymerization(GO:1901880) |
| 0.0 | 1.3 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.6 | GO:0006541 | glutamine metabolic process(GO:0006541) |
| 0.0 | 0.8 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.6 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.0 | 0.6 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 1.7 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 0.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.3 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.2 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.5 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 1.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 2.0 | GO:1901342 | regulation of vasculature development(GO:1901342) |
| 0.0 | 1.8 | GO:0007224 | smoothened signaling pathway(GO:0007224) |
| 0.0 | 5.8 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 0.2 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 2.0 | GO:0070507 | regulation of microtubule cytoskeleton organization(GO:0070507) |
| 0.0 | 1.5 | GO:0071466 | cellular response to xenobiotic stimulus(GO:0071466) |
| 0.0 | 0.9 | GO:0035272 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 0.2 | GO:0046627 | regulation of insulin receptor signaling pathway(GO:0046626) negative regulation of insulin receptor signaling pathway(GO:0046627) regulation of cellular response to insulin stimulus(GO:1900076) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 1.5 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 0.1 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 1.0 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 0.7 | GO:0051056 | regulation of small GTPase mediated signal transduction(GO:0051056) |
| 0.0 | 0.5 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.6 | GO:0016358 | dendrite development(GO:0016358) |
| 0.0 | 0.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.3 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.7 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.3 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 3.5 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.0 | 0.3 | GO:0070534 | protein K63-linked ubiquitination(GO:0070534) |
| 0.0 | 0.7 | GO:0009247 | glycolipid biosynthetic process(GO:0009247) |
| 0.0 | 0.7 | GO:0009416 | response to light stimulus(GO:0009416) |
| 0.0 | 0.9 | GO:0045055 | regulated exocytosis(GO:0045055) |
| 0.0 | 0.6 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.3 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 2.4 | 11.8 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 2.0 | 12.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 1.3 | 7.6 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.9 | 6.0 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.7 | 3.0 | GO:0016586 | RSC complex(GO:0016586) |
| 0.7 | 2.2 | GO:0071914 | prominosome(GO:0071914) |
| 0.7 | 12.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.5 | 11.5 | GO:0005861 | troponin complex(GO:0005861) |
| 0.3 | 1.3 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.3 | 1.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.3 | 1.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.3 | 2.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.2 | 1.9 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 1.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 2.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 2.9 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.2 | 0.8 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.2 | 0.8 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 0.8 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.2 | 3.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 0.6 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.1 | 1.0 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 2.9 | GO:0005940 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 1.8 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.6 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 5.3 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 12.1 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.1 | 1.1 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 2.1 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.1 | 0.6 | GO:0000803 | sex chromosome(GO:0000803) inactive sex chromosome(GO:0098577) |
| 0.1 | 5.3 | GO:0044420 | extracellular matrix component(GO:0044420) |
| 0.1 | 3.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.1 | 0.6 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.1 | 1.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 1.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 3.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.6 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.0 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 2.8 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 1.1 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 0.6 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 3.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.1 | 2.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 3.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 3.1 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 4.9 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 3.5 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 1.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 1.9 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.9 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 0.8 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.1 | 1.7 | GO:0030424 | axon(GO:0030424) |
| 0.1 | 1.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 3.1 | GO:0005684 | U2-type spliceosomal complex(GO:0005684) |
| 0.0 | 1.0 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 3.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.8 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 2.3 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 3.9 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.9 | GO:0005911 | cell-cell junction(GO:0005911) |
| 0.0 | 0.3 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 8.7 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.4 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 1.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 3.5 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.0 | 0.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 1.2 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.2 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 20.6 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 45.5 | GO:0005886 | plasma membrane(GO:0005886) |
| 0.0 | 0.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 2.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 1.8 | GO:0045202 | synapse(GO:0045202) |
| 0.0 | 0.6 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 1.1 | GO:0030659 | vesicle membrane(GO:0012506) cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 11.8 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 2.2 | 8.9 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 1.9 | 5.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 1.9 | 13.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 1.6 | 6.3 | GO:0070325 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 1.4 | 5.5 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 1.1 | 4.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.1 | 3.2 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 1.0 | 3.9 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.9 | 26.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.9 | 4.5 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.8 | 3.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.8 | 3.0 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.7 | 11.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.7 | 7.2 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.7 | 3.5 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.7 | 2.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.7 | 10.1 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.7 | 7.2 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.6 | 1.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.6 | 4.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.5 | 2.7 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.5 | 3.9 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.5 | 1.8 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.4 | 2.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 3.5 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.4 | 1.1 | GO:0048019 | receptor inhibitor activity(GO:0030547) co-receptor binding(GO:0039706) receptor antagonist activity(GO:0048019) |
| 0.3 | 3.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.3 | 3.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.3 | 8.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.3 | 1.1 | GO:0099528 | G-protein coupled neurotransmitter receptor activity(GO:0099528) |
| 0.3 | 1.9 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.3 | 0.8 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.3 | 3.6 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.3 | 1.0 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.2 | 3.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 6.0 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 8.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.2 | 2.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 0.9 | GO:0019809 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.2 | 3.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.2 | 1.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 1.9 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.2 | 2.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 0.6 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.2 | 3.5 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.2 | 0.8 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.2 | 2.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 3.7 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.2 | 0.6 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.2 | 2.0 | GO:0022840 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.2 | 10.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 1.8 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.2 | 0.5 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.2 | 0.8 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.2 | 1.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 3.1 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.2 | 3.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.2 | 7.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.2 | 0.9 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.2 | 1.9 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.2 | 1.8 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.6 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 2.5 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.4 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 1.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 2.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 3.4 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 1.0 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 0.6 | GO:0001097 | TFIIH-class transcription factor binding(GO:0001097) poly(C) RNA binding(GO:0017130) |
| 0.1 | 1.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 1.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.1 | 4.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 2.8 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 0.8 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.8 | GO:0052812 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 0.6 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 0.5 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.1 | 4.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.3 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 37.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 1.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.1 | 3.2 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.1 | 0.2 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.1 | 3.7 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 0.9 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.1 | 12.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 1.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.8 | GO:0019201 | nucleotide kinase activity(GO:0019201) |
| 0.0 | 1.0 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 9.6 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 4.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 3.8 | GO:0031406 | carboxylic acid binding(GO:0031406) |
| 0.0 | 19.5 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.6 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 1.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 33.7 | GO:0001159 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 1.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 2.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 2.5 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 6.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.7 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 1.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 7.6 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.8 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.6 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.9 | GO:0030674 | protein binding, bridging(GO:0030674) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.8 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 1.0 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 2.0 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.9 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.6 | GO:0016879 | ligase activity, forming carbon-nitrogen bonds(GO:0016879) |
| 0.0 | 0.7 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.5 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.4 | 3.5 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 5.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 3.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 4.5 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 1.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 3.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 6.5 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 1.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 2.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.1 | 2.5 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 2.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 2.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.0 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 2.9 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.7 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 12.5 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.8 | 12.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.6 | 3.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.4 | 5.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.4 | 3.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.4 | 2.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.4 | 5.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.3 | 1.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.2 | 2.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.2 | 4.0 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.2 | 3.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 2.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.2 | 1.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 1.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 0.8 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 2.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.1 | 1.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 1.0 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 2.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.5 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 2.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.8 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 1.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |