Project
DANIO-CODE
Navigation
Downloads
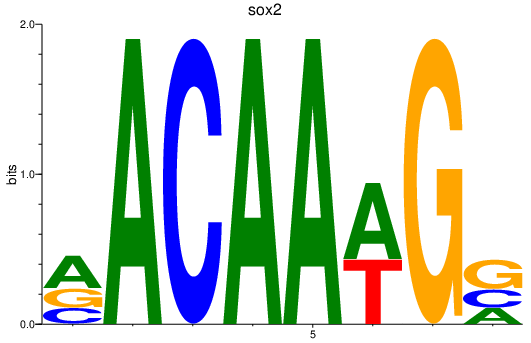
Results for sox2
Z-value: 2.37
Transcription factors associated with sox2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox2
|
ENSDARG00000070913 | SRY-box transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox2 | dr10_dc_chr22_-_37414940_37415112 | 0.86 | 2.0e-05 | Click! |
Activity profile of sox2 motif
Sorted Z-values of sox2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of sox2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_39173765 | 7.11 |
ENSDART00000173748
|
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr14_+_9115745 | 6.59 |
ENSDART00000123652
|
hmgn6
|
high mobility group nucleosome binding domain 6 |
| chr14_-_7774262 | 6.49 |
ENSDART00000045109
|
zgc:92242
|
zgc:92242 |
| chr10_+_33227967 | 5.54 |
ENSDART00000159666
|
myl10
|
myosin, light chain 10, regulatory |
| chr19_+_41396014 | 5.38 |
ENSDART00000159444
ENSDART00000042990 ENSDART00000144544 |
col1a2
|
collagen, type I, alpha 2 |
| chr1_-_44009572 | 5.21 |
ENSDART00000144900
|
ENSDARG00000079632
|
ENSDARG00000079632 |
| chr1_+_16683931 | 4.89 |
ENSDART00000103262
ENSDART00000145068 ENSDART00000169619 ENSDART00000010526 |
fat1a
|
FAT atypical cadherin 1a |
| chr8_-_16662185 | 4.88 |
ENSDART00000076542
|
rpe65b
|
retinal pigment epithelium-specific protein 65b |
| chr6_-_43094573 | 4.71 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr7_-_16776026 | 4.63 |
ENSDART00000022441
|
dbx1a
|
developing brain homeobox 1a |
| chr12_-_30662709 | 4.45 |
ENSDART00000126466
|
entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr23_-_7892862 | 4.44 |
ENSDART00000157612
ENSDART00000165427 |
myt1b
|
myelin transcription factor 1b |
| chr21_+_26352675 | 4.20 |
ENSDART00000077197
|
tmsb
|
thymosin, beta |
| chr23_+_35609887 | 4.19 |
ENSDART00000179393
|
tuba1b
|
tubulin, alpha 1b |
| chr7_+_44373815 | 4.13 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr20_+_31366832 | 4.10 |
ENSDART00000133353
|
apobb.1
|
apolipoprotein Bb, tandem duplicate 1 |
| chr4_-_24310846 | 4.02 |
ENSDART00000017443
|
celf2
|
cugbp, Elav-like family member 2 |
| chr2_-_43110857 | 4.00 |
ENSDART00000098303
|
oc90
|
otoconin 90 |
| chr10_+_44195410 | 3.92 |
ENSDART00000046172
|
cryba4
|
crystallin, beta A4 |
| chr3_+_28450576 | 3.90 |
ENSDART00000150893
|
sept12
|
septin 12 |
| chr1_-_48853800 | 3.82 |
ENSDART00000137357
|
zgc:175214
|
zgc:175214 |
| chr24_-_33869817 | 3.78 |
ENSDART00000079283
|
tmeff1b
|
transmembrane protein with EGF-like and two follistatin-like domains 1b |
| chr3_-_26393945 | 3.71 |
ENSDART00000087118
|
xylt1
|
xylosyltransferase I |
| chr14_-_36037883 | 3.70 |
ENSDART00000173006
|
gpm6aa
|
glycoprotein M6Aa |
| chr13_+_22545318 | 3.62 |
ENSDART00000143312
|
zgc:193505
|
zgc:193505 |
| chr16_+_40610589 | 3.50 |
ENSDART00000038294
|
tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr24_-_37680649 | 3.48 |
ENSDART00000056286
|
h1f0
|
H1 histone family, member 0 |
| chr6_+_27156169 | 3.47 |
ENSDART00000088364
|
kif1aa
|
kinesin family member 1Aa |
| chr6_+_47845002 | 3.46 |
ENSDART00000140943
|
padi2
|
peptidyl arginine deiminase, type II |
| chr13_-_18506153 | 3.45 |
ENSDART00000057869
|
mat1a
|
methionine adenosyltransferase I, alpha |
| chr16_+_4825019 | 3.45 |
ENSDART00000167665
|
LIN28A
|
lin-28 homolog A |
| chr19_+_7234029 | 3.44 |
ENSDART00000080348
|
brd2a
|
bromodomain containing 2a |
| chr7_-_7164950 | 3.38 |
ENSDART00000012637
|
zgc:101810
|
zgc:101810 |
| chr3_-_54893038 | 3.38 |
ENSDART00000155871
ENSDART00000109016 |
hbae3
|
hemoglobin alpha embryonic-3 |
| chr13_-_13163676 | 3.37 |
ENSDART00000125883
ENSDART00000013534 |
fgfr3
|
fibroblast growth factor receptor 3 |
| chr10_+_18994733 | 3.35 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr13_-_869704 | 3.35 |
|
|
|
| chr11_-_29891067 | 3.30 |
ENSDART00000172106
|
scml2
|
sex comb on midleg-like 2 (Drosophila) |
| chr3_-_25683205 | 3.27 |
ENSDART00000169706
|
ntn1b
|
netrin 1b |
| chr2_-_30675594 | 3.27 |
ENSDART00000087270
|
ctnnd2b
|
catenin (cadherin-associated protein), delta 2b |
| chr16_-_46799406 | 3.26 |
ENSDART00000115244
|
MEX3A
|
mex-3 RNA binding family member A |
| chr14_+_38445969 | 3.26 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr16_-_24697750 | 3.25 |
ENSDART00000163305
|
fxyd6l
|
FXYD domain containing ion transport regulator 6 like |
| chr10_+_22759607 | 3.22 |
|
|
|
| chr24_+_24924379 | 3.08 |
ENSDART00000115165
|
amer2
|
APC membrane recruitment protein 2 |
| chr21_-_35806638 | 3.05 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr19_+_5399813 | 3.05 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr25_-_14472107 | 3.04 |
|
|
|
| chr17_-_36988937 | 3.04 |
ENSDART00000145236
|
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr16_+_1302842 | 3.04 |
|
|
|
| chr11_+_3186396 | 3.02 |
ENSDART00000159459
|
pmela
|
premelanosome protein a |
| chr11_+_6118409 | 3.01 |
ENSDART00000027666
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr13_-_31310439 | 2.98 |
ENSDART00000076571
|
rtn1a
|
reticulon 1a |
| chr14_+_23980348 | 2.97 |
ENSDART00000160984
|
BX005228.1
|
ENSDARG00000105465 |
| chr9_-_42894582 | 2.97 |
ENSDART00000144744
|
col5a2a
|
collagen, type V, alpha 2a |
| chr22_+_17091411 | 2.93 |
ENSDART00000170076
|
frem1b
|
Fras1 related extracellular matrix 1b |
| chr23_+_35819625 | 2.92 |
ENSDART00000049551
|
rarga
|
retinoic acid receptor gamma a |
| chr11_-_6004509 | 2.90 |
ENSDART00000108628
|
ano8b
|
anoctamin 8b |
| chr9_+_30279907 | 2.88 |
ENSDART00000102981
|
col8a1a
|
collagen, type VIII, alpha 1a |
| chr4_-_12863235 | 2.87 |
ENSDART00000080536
|
hmga2
|
high mobility group AT-hook 2 |
| chr23_+_27141681 | 2.86 |
ENSDART00000054238
|
mipa
|
major intrinsic protein of lens fiber a |
| chr16_+_25401922 | 2.82 |
ENSDART00000172514
ENSDART00000086409 |
dync1i1
|
dynein, cytoplasmic 1, intermediate chain 1 |
| chr15_+_46799979 | 2.82 |
|
|
|
| chr4_-_11582061 | 2.81 |
ENSDART00000049066
|
net1
|
neuroepithelial cell transforming 1 |
| chr13_+_25298383 | 2.79 |
|
|
|
| chr11_-_29910947 | 2.78 |
ENSDART00000156121
|
scml2
|
sex comb on midleg-like 2 (Drosophila) |
| chr14_-_40918015 | 2.78 |
ENSDART00000147389
|
tmem35
|
transmembrane protein 35 |
| chr8_-_38168395 | 2.77 |
ENSDART00000155189
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr21_-_37286942 | 2.74 |
ENSDART00000100310
|
dbn1
|
drebrin 1 |
| chr13_+_1051015 | 2.73 |
ENSDART00000033528
|
tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr9_+_38975508 | 2.72 |
|
|
|
| chr2_-_8219329 | 2.72 |
ENSDART00000040209
|
ephb3a
|
eph receptor B3a |
| chr23_-_30119058 | 2.71 |
ENSDART00000103480
|
ccdc187
|
coiled-coil domain containing 187 |
| chr4_+_3344998 | 2.71 |
ENSDART00000075320
|
nampta
|
nicotinamide phosphoribosyltransferase a |
| chr23_+_32573474 | 2.66 |
ENSDART00000134811
|
si:dkey-261h17.1
|
si:dkey-261h17.1 |
| chr22_-_22846670 | 2.66 |
ENSDART00000176355
|
CABZ01039424.2
|
ENSDARG00000108442 |
| chr5_-_31692803 | 2.66 |
ENSDART00000131983
|
myhz1.2
|
myosin, heavy polypeptide 1.2, skeletal muscle |
| chr2_+_26523457 | 2.65 |
ENSDART00000024662
|
plppr3a
|
phospholipid phosphatase related 3a |
| chr15_+_28435937 | 2.65 |
ENSDART00000142298
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr6_-_11544518 | 2.64 |
ENSDART00000151195
|
march7
|
membrane-associated ring finger (C3HC4) 7 |
| chr10_-_17630376 | 2.64 |
ENSDART00000113101
|
smarcad1b
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 b |
| chr13_-_39033893 | 2.63 |
ENSDART00000045434
|
col9a1b
|
collagen, type IX, alpha 1b |
| chr5_-_30324182 | 2.62 |
ENSDART00000153909
|
spns2
|
spinster homolog 2 (Drosophila) |
| chr3_+_23567458 | 2.62 |
ENSDART00000078466
|
hoxb3a
|
homeobox B3a |
| chr13_-_33137075 | 2.61 |
ENSDART00000138096
|
oip5-as1
|
oip5 antisense RNA 1 |
| chr2_-_50638153 | 2.60 |
ENSDART00000108900
|
cntnap2b
|
contactin associated protein-like 2b |
| chr9_-_22371512 | 2.59 |
ENSDART00000101809
|
crygm2d6
|
crystallin, gamma M2d6 |
| chr7_-_24201833 | 2.57 |
ENSDART00000121684
|
nat8l
|
N-acetyltransferase 8-like |
| chr6_-_43094926 | 2.55 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr2_+_48448974 | 2.55 |
ENSDART00000023040
|
hes6
|
hes family bHLH transcription factor 6 |
| chr18_-_6494014 | 2.51 |
ENSDART00000062423
|
tnni1c
|
troponin I, skeletal, slow c |
| chr5_-_66792947 | 2.51 |
ENSDART00000147009
|
si:dkey-251i10.2
|
si:dkey-251i10.2 |
| chr10_-_27261937 | 2.50 |
|
|
|
| chr9_+_36092350 | 2.50 |
ENSDART00000005086
|
atp1a1b
|
ATPase, Na+/K+ transporting, alpha 1b polypeptide |
| chr10_-_36690119 | 2.49 |
ENSDART00000077161
|
rsf1b.1
|
remodeling and spacing factor 1b, tandem duplicate 1 |
| chr16_+_33639132 | 2.48 |
ENSDART00000013148
|
pou3f1
|
POU class 3 homeobox 1 |
| chr25_+_19992389 | 2.48 |
ENSDART00000143441
|
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr5_-_30015572 | 2.48 |
ENSDART00000147769
|
si:ch211-117m20.5
|
si:ch211-117m20.5 |
| chr20_-_20412830 | 2.47 |
ENSDART00000114779
|
ENSDARG00000079369
|
ENSDARG00000079369 |
| chr19_+_12995955 | 2.47 |
ENSDART00000132892
|
cthrc1a
|
collagen triple helix repeat containing 1a |
| chr14_-_36523075 | 2.45 |
ENSDART00000109293
|
rnf130
|
ring finger protein 130 |
| chr17_+_38528709 | 2.44 |
ENSDART00000123298
|
stard9
|
StAR-related lipid transfer (START) domain containing 9 |
| chr13_-_5440923 | 2.44 |
ENSDART00000102576
|
meis1b
|
Meis homeobox 1 b |
| chr19_-_7531709 | 2.44 |
ENSDART00000104750
|
mllt11
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 11 |
| chr2_+_38178934 | 2.44 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr20_-_48677794 | 2.44 |
ENSDART00000124040
ENSDART00000148437 |
insm1a
|
insulinoma-associated 1a |
| chr10_+_9716807 | 2.44 |
ENSDART00000064977
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr16_+_24697776 | 2.42 |
|
|
|
| chr21_-_11539798 | 2.41 |
ENSDART00000144770
|
cast
|
calpastatin |
| chr11_-_42933969 | 2.39 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr4_-_12862869 | 2.39 |
ENSDART00000080536
|
hmga2
|
high mobility group AT-hook 2 |
| chr16_+_46327528 | 2.39 |
ENSDART00000040769
|
nr2f5
|
nuclear receptor subfamily 2, group F, member 5 |
| chr20_+_53310252 | 2.38 |
ENSDART00000175125
|
CABZ01089116.1
|
ENSDARG00000106770 |
| chr4_-_25075693 | 2.36 |
ENSDART00000025153
|
gata3
|
GATA binding protein 3 |
| chr6_+_7309023 | 2.35 |
ENSDART00000160128
|
erbb3a
|
erb-b2 receptor tyrosine kinase 3a |
| chr25_+_31457309 | 2.35 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr4_-_12862708 | 2.34 |
ENSDART00000080536
|
hmga2
|
high mobility group AT-hook 2 |
| chr11_-_26595578 | 2.33 |
ENSDART00000153519
|
iqsec1b
|
IQ motif and Sec7 domain 1b |
| chr6_+_6967174 | 2.32 |
ENSDART00000151311
|
ENSDARG00000058903
|
ENSDARG00000058903 |
| chr7_-_32888309 | 2.32 |
ENSDART00000173461
|
BX571811.2
|
ENSDARG00000105655 |
| chr18_+_22617072 | 2.30 |
ENSDART00000128965
|
bcar1
|
breast cancer anti-estrogen resistance 1 |
| chr17_-_6219019 | 2.30 |
|
|
|
| chr1_-_31123999 | 2.30 |
ENSDART00000152493
|
nt5c2b
|
5'-nucleotidase, cytosolic IIb |
| chr2_+_20118917 | 2.29 |
ENSDART00000038648
|
ptbp2b
|
polypyrimidine tract binding protein 2b |
| chr9_-_22318918 | 2.28 |
ENSDART00000124272
|
crygm2d17
|
crystallin, gamma M2d17 |
| chr16_-_22797736 | 2.27 |
ENSDART00000143836
|
si:ch211-105c13.3
|
si:ch211-105c13.3 |
| chr1_-_49004615 | 2.26 |
ENSDART00000134399
|
slkb
|
STE20-like kinase b |
| chr3_+_26896869 | 2.26 |
ENSDART00000065495
|
emp2
|
epithelial membrane protein 2 |
| chr17_+_23278879 | 2.26 |
ENSDART00000153652
|
zgc:165461
|
zgc:165461 |
| chr15_-_31687 | 2.25 |
|
|
|
| chr3_+_54327353 | 2.24 |
ENSDART00000127487
|
eif3g
|
eukaryotic translation initiation factor 3, subunit G |
| chr5_+_8415025 | 2.24 |
ENSDART00000046440
|
agpat9l
|
1-acylglycerol-3-phosphate O-acyltransferase 9, like |
| chr25_+_18866796 | 2.23 |
ENSDART00000017299
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr21_-_1716495 | 2.22 |
ENSDART00000151049
|
onecut2
|
one cut homeobox 2 |
| chr16_+_5778650 | 2.21 |
ENSDART00000131575
|
zgc:158689
|
zgc:158689 |
| chr13_-_13163801 | 2.21 |
ENSDART00000156968
|
fgfr3
|
fibroblast growth factor receptor 3 |
| chr24_+_389982 | 2.20 |
ENSDART00000061973
|
tgfbr1b
|
transforming growth factor, beta receptor 1 b |
| chr25_+_30746445 | 2.19 |
ENSDART00000156916
|
lsp1
|
lymphocyte-specific protein 1 |
| chr23_+_6043862 | 2.19 |
|
|
|
| chr14_-_26406720 | 2.19 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr15_-_19836573 | 2.18 |
ENSDART00000114888
|
picalmb
|
phosphatidylinositol binding clathrin assembly protein b |
| chr6_-_50705420 | 2.18 |
ENSDART00000074100
|
osgn1
|
oxidative stress induced growth inhibitor 1 |
| chr14_+_151136 | 2.17 |
ENSDART00000165766
|
mcm7
|
minichromosome maintenance complex component 7 |
| chr15_+_23849554 | 2.15 |
ENSDART00000138375
|
ift20
|
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr10_+_4717800 | 2.14 |
ENSDART00000161789
|
palm2
|
paralemmin 2 |
| chr13_+_21989327 | 2.13 |
ENSDART00000173206
|
camk2g2
|
calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma 2 |
| chr24_-_40968409 | 2.13 |
ENSDART00000169315
ENSDART00000171543 |
smyhc1
|
slow myosin heavy chain 1 |
| chr16_+_5778398 | 2.12 |
ENSDART00000011166
ENSDART00000126445 |
zgc:158689
|
zgc:158689 |
| chr9_+_42112576 | 2.11 |
ENSDART00000130434
ENSDART00000007058 |
col18a1a
|
collagen type XVIII alpha 1 chain a |
| chr13_+_24370456 | 2.11 |
ENSDART00000077899
|
sertad2b
|
SERTA domain containing 2b |
| chr23_+_25428372 | 2.11 |
ENSDART00000147440
|
fmnl3
|
formin-like 3 |
| chr23_+_23730717 | 2.10 |
|
|
|
| chr14_-_30556788 | 2.10 |
ENSDART00000087918
|
slc7a3b
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 3b |
| chr20_+_54512601 | 2.09 |
ENSDART00000169386
|
faua
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed a |
| chr6_+_7936632 | 2.09 |
ENSDART00000146106
|
nfil3-5
|
nuclear factor, interleukin 3 regulated, member 5 |
| chr2_+_55859099 | 2.09 |
ENSDART00000097753
ENSDART00000141688 |
nmrk2
|
nicotinamide riboside kinase 2 |
| chr11_-_42263721 | 2.08 |
ENSDART00000130573
|
atp6ap1la
|
ATPase, H+ transporting, lysosomal accessory protein 1-like a |
| chr5_+_19802818 | 2.08 |
|
|
|
| chr18_-_25010085 | 2.08 |
ENSDART00000133786
|
si:ch211-196l7.4
|
si:ch211-196l7.4 |
| chr8_-_12171624 | 2.08 |
ENSDART00000132824
|
dab2ipa
|
DAB2 interacting protein a |
| chr16_+_20393077 | 2.07 |
|
|
|
| chr20_+_4567653 | 2.06 |
|
|
|
| chr23_+_44817648 | 2.06 |
ENSDART00000143688
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr21_+_25034997 | 2.06 |
ENSDART00000167523
|
dixdc1b
|
DIX domain containing 1b |
| chr3_-_30027552 | 2.05 |
ENSDART00000103502
|
si:ch211-152f23.5
|
si:ch211-152f23.5 |
| chr8_+_15968469 | 2.05 |
ENSDART00000134787
|
elavl4
|
ELAV like neuron-specific RNA binding protein 4 |
| chr10_+_26705729 | 2.03 |
ENSDART00000147013
|
si:ch73-52f15.5
|
si:ch73-52f15.5 |
| chr18_+_44802349 | 2.03 |
ENSDART00000139526
|
fam118b
|
family with sequence similarity 118, member B |
| chr24_+_13780640 | 2.01 |
ENSDART00000136443
ENSDART00000081595 ENSDART00000012253 |
eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr4_+_1667305 | 1.99 |
ENSDART00000150198
|
slc38a4
|
solute carrier family 38, member 4 |
| chr14_-_9114872 | 1.99 |
ENSDART00000081158
|
sh3bgrl
|
SH3 domain binding glutamate-rich protein like |
| chr3_-_35928594 | 1.98 |
|
|
|
| chr21_-_24552672 | 1.98 |
|
|
|
| chr6_+_6767424 | 1.96 |
ENSDART00000065551
ENSDART00000151393 |
zak
|
sterile alpha motif and leucine zipper containing kinase AZK |
| chr5_+_49103778 | 1.95 |
ENSDART00000155405
|
CR751602.2
|
ENSDARG00000098045 |
| chr17_+_45594103 | 1.95 |
ENSDART00000153922
|
si:ch211-202f3.3
|
si:ch211-202f3.3 |
| chr4_-_73484546 | 1.94 |
|
|
|
| chr18_-_36291570 | 1.94 |
|
|
|
| chr23_-_42775849 | 1.94 |
ENSDART00000149944
|
gpx7
|
glutathione peroxidase 7 |
| chr3_-_54959360 | 1.94 |
ENSDART00000154031
|
kank2
|
KN motif and ankyrin repeat domains 2 |
| chr7_+_48532749 | 1.94 |
ENSDART00000145375
|
cpt1aa
|
carnitine palmitoyltransferase 1Aa (liver) |
| chr23_+_24141576 | 1.94 |
|
|
|
| chr12_-_9400344 | 1.94 |
ENSDART00000003932
|
erbb2
|
erb-b2 receptor tyrosine kinase 2 |
| chr21_-_19280266 | 1.93 |
ENSDART00000141596
|
gpat3
|
glycerol-3-phosphate acyltransferase 3 |
| chr9_-_12840621 | 1.93 |
ENSDART00000088042
|
myo10l3
|
myosin X-like 3 |
| chr22_-_33942334 | 1.93 |
|
|
|
| chr21_-_25704662 | 1.92 |
ENSDART00000101211
|
cldnh
|
claudin h |
| chr1_+_25662910 | 1.92 |
ENSDART00000113020
|
tet2
|
tet methylcytosine dioxygenase 2 |
| chr15_-_4537178 | 1.92 |
ENSDART00000155619
ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr9_+_4188263 | 1.92 |
ENSDART00000164543
|
kalrna
|
kalirin RhoGEF kinase a |
| chr5_-_14148028 | 1.92 |
ENSDART00000113037
|
sema4c
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C |
| chr5_+_2384929 | 1.92 |
|
|
|
| KN149932v1_+_27584 | 1.92 |
|
|
|
| chr1_+_53353690 | 1.91 |
ENSDART00000126339
|
dla
|
deltaA |
| chr11_+_23522743 | 1.91 |
ENSDART00000121874
|
nfasca
|
neurofascin homolog (chicken) a |
| chr13_+_27102308 | 1.91 |
ENSDART00000145901
|
rin2
|
Ras and Rab interactor 2 |
| chr19_-_3299610 | 1.91 |
ENSDART00000105168
|
si:ch211-133n4.4
|
si:ch211-133n4.4 |
| chr16_-_38117896 | 1.91 |
ENSDART00000114266
|
plekho1b
|
pleckstrin homology domain containing, family O member 1b |
| chr11_+_26371444 | 1.91 |
ENSDART00000042322
|
map1lc3a
|
microtubule-associated protein 1 light chain 3 alpha |
| chr9_-_23081918 | 1.90 |
ENSDART00000143888
|
neb
|
nebulin |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.1 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 1.4 | 4.2 | GO:0019730 | antimicrobial humoral response(GO:0019730) antibacterial humoral response(GO:0019731) |
| 1.1 | 4.6 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 1.0 | 4.2 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 1.0 | 7.3 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 1.0 | 3.0 | GO:0097435 | fibril organization(GO:0097435) |
| 0.9 | 2.7 | GO:0035523 | protein K29-linked deubiquitination(GO:0035523) protein K33-linked deubiquitination(GO:1990168) |
| 0.9 | 4.3 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 0.8 | 3.4 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.8 | 4.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.8 | 4.0 | GO:0050482 | icosanoid secretion(GO:0032309) arachidonic acid secretion(GO:0050482) arachidonate transport(GO:1903963) |
| 0.7 | 3.7 | GO:0006211 | 5-methylcytosine catabolic process(GO:0006211) 5-methylcytosine metabolic process(GO:0019857) |
| 0.7 | 4.4 | GO:0033986 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.7 | 3.7 | GO:0051899 | membrane depolarization(GO:0051899) |
| 0.7 | 8.7 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.7 | 2.1 | GO:0035790 | regulation of platelet-derived growth factor receptor signaling pathway(GO:0010640) platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) |
| 0.7 | 2.1 | GO:1903352 | ornithine transport(GO:0015822) L-ornithine transmembrane transport(GO:1903352) |
| 0.7 | 2.1 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) |
| 0.7 | 2.6 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.7 | 3.3 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.7 | 2.0 | GO:0048940 | anterior lateral line nerve development(GO:0048909) anterior lateral line nerve glial cell differentiation(GO:0048913) myelination of anterior lateral line nerve axons(GO:0048914) anterior lateral line nerve glial cell development(GO:0048939) anterior lateral line nerve glial cell morphogenesis involved in differentiation(GO:0048940) |
| 0.6 | 1.9 | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) cellular response to vitamin D(GO:0071305) |
| 0.6 | 2.4 | GO:0003210 | cardiac atrium formation(GO:0003210) |
| 0.6 | 1.8 | GO:1903441 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to ciliary membrane(GO:1903441) |
| 0.6 | 6.4 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.6 | 1.7 | GO:0072025 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 0.6 | 0.6 | GO:0046717 | acid secretion(GO:0046717) |
| 0.6 | 1.7 | GO:0048903 | anterior lateral line neuromast development(GO:0048901) anterior lateral line neuromast hair cell differentiation(GO:0048903) |
| 0.6 | 1.7 | GO:0072526 | pyridine nucleotide catabolic process(GO:0019364) NAD catabolic process(GO:0019677) pyridine-containing compound catabolic process(GO:0072526) |
| 0.5 | 1.6 | GO:0050810 | regulation of steroid biosynthetic process(GO:0050810) |
| 0.5 | 2.0 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.5 | 2.0 | GO:0050687 | negative regulation of response to biotic stimulus(GO:0002832) negative regulation of defense response to virus(GO:0050687) |
| 0.5 | 3.0 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.5 | 3.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.5 | 1.9 | GO:0055016 | hypochord development(GO:0055016) |
| 0.5 | 1.4 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.5 | 1.9 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.5 | 1.4 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.5 | 1.4 | GO:0001659 | temperature homeostasis(GO:0001659) sleep(GO:0030431) |
| 0.5 | 1.4 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.5 | 1.9 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.5 | 1.8 | GO:0048025 | negative regulation of RNA splicing(GO:0033119) negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.5 | 1.8 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.4 | 1.3 | GO:0061379 | associative learning(GO:0008306) visual learning(GO:0008542) mammillary axonal complex development(GO:0061373) mammillothalamic axonal tract development(GO:0061374) corpora quadrigemina development(GO:0061378) inferior colliculus development(GO:0061379) cell migration in diencephalon(GO:0061381) |
| 0.4 | 1.8 | GO:1901998 | toxin transport(GO:1901998) |
| 0.4 | 8.0 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.4 | 1.3 | GO:0002706 | regulation of lymphocyte mediated immunity(GO:0002706) |
| 0.4 | 3.5 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.4 | 5.7 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.4 | 1.3 | GO:1900271 | regulation of presynaptic cytosolic calcium ion concentration(GO:0099509) regulation of long-term synaptic potentiation(GO:1900271) |
| 0.4 | 0.9 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.4 | 1.3 | GO:1903010 | regulation of bone development(GO:1903010) |
| 0.4 | 1.7 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.4 | 4.2 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.4 | 3.7 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.4 | 2.9 | GO:0006833 | water transport(GO:0006833) |
| 0.4 | 1.6 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.4 | 3.2 | GO:2000251 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.4 | 3.6 | GO:0097340 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.4 | 1.6 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.4 | 10.8 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.4 | 1.5 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.4 | 3.8 | GO:0007271 | synaptic transmission, cholinergic(GO:0007271) |
| 0.4 | 1.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.4 | 1.9 | GO:1901739 | regulation of myoblast fusion(GO:1901739) |
| 0.4 | 4.2 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.4 | 3.0 | GO:0014831 | gastro-intestinal system smooth muscle contraction(GO:0014831) |
| 0.4 | 0.7 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.4 | 1.1 | GO:1902571 | regulation of serine-type peptidase activity(GO:1902571) |
| 0.4 | 5.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.4 | 1.1 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.4 | 2.8 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.3 | 1.0 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) antigen processing and presentation of exogenous antigen(GO:0019884) antigen processing and presentation of exogenous peptide antigen via MHC class I(GO:0042590) |
| 0.3 | 1.0 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.3 | 1.4 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.3 | 2.1 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.3 | 6.2 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.3 | 5.8 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.3 | 2.0 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.3 | 1.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.3 | 0.3 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.3 | 2.3 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.3 | 3.5 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.3 | 4.2 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.3 | 1.0 | GO:0051482 | negative regulation of adenylate cyclase activity(GO:0007194) negative regulation of cyclic nucleotide metabolic process(GO:0030800) negative regulation of cyclic nucleotide biosynthetic process(GO:0030803) negative regulation of nucleotide biosynthetic process(GO:0030809) negative regulation of cAMP metabolic process(GO:0030815) negative regulation of cAMP biosynthetic process(GO:0030818) negative regulation of cyclase activity(GO:0031280) negative regulation of lyase activity(GO:0051350) positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) negative regulation of purine nucleotide biosynthetic process(GO:1900372) |
| 0.3 | 1.6 | GO:0042985 | negative regulation of glycoprotein biosynthetic process(GO:0010561) negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) negative regulation of glycoprotein metabolic process(GO:1903019) |
| 0.3 | 4.1 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.3 | 1.3 | GO:0048677 | axon extension involved in regeneration(GO:0048677) sprouting of injured axon(GO:0048682) |
| 0.3 | 0.6 | GO:0061011 | hepatic duct development(GO:0061011) |
| 0.3 | 2.5 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.3 | 0.9 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.3 | 3.4 | GO:0015669 | gas transport(GO:0015669) oxygen transport(GO:0015671) |
| 0.3 | 2.1 | GO:0060581 | ventral spinal cord interneuron fate commitment(GO:0060579) cell fate commitment involved in pattern specification(GO:0060581) |
| 0.3 | 1.8 | GO:0042438 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.3 | 1.8 | GO:0060829 | negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.3 | 0.9 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.3 | 1.5 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.3 | 3.6 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.3 | 4.4 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.3 | 1.1 | GO:0032048 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.3 | 0.9 | GO:1990359 | detoxification of zinc ion(GO:0010312) stress response to zinc ion(GO:1990359) |
| 0.3 | 0.9 | GO:1902895 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.3 | 4.0 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.3 | 1.7 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.3 | 2.5 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.3 | 2.0 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.3 | 1.7 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 0.3 | 1.6 | GO:0048923 | posterior lateral line neuromast hair cell differentiation(GO:0048923) |
| 0.3 | 1.9 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.3 | 0.8 | GO:0019284 | L-methionine biosynthetic process from S-adenosylmethionine(GO:0019284) |
| 0.3 | 1.3 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) positive regulation of cartilage development(GO:0061036) |
| 0.3 | 0.8 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.3 | 2.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.2 | 5.4 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.2 | 1.7 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.2 | 2.2 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.2 | 2.9 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.2 | 16.2 | GO:0007601 | visual perception(GO:0007601) |
| 0.2 | 1.4 | GO:2001240 | histone dephosphorylation(GO:0016576) negative regulation of signal transduction in absence of ligand(GO:1901099) regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001239) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.2 | 0.7 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.2 | 1.4 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.2 | 1.7 | GO:1902915 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 | 1.2 | GO:0002446 | neutrophil mediated immunity(GO:0002446) defense response to fungus(GO:0050832) |
| 0.2 | 0.7 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.2 | 5.1 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.2 | 1.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.2 | 1.4 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.2 | 2.2 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.2 | 2.7 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.2 | 2.9 | GO:0014028 | notochord formation(GO:0014028) |
| 0.2 | 1.1 | GO:0060416 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.2 | 3.3 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.2 | 0.6 | GO:0051701 | interaction with host(GO:0051701) |
| 0.2 | 0.9 | GO:0050936 | xanthophore differentiation(GO:0050936) |
| 0.2 | 0.6 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 1.2 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.2 | 4.1 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.2 | 1.4 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.2 | 1.4 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.2 | 1.2 | GO:0036268 | swimming(GO:0036268) |
| 0.2 | 1.5 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.2 | 1.3 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.2 | 0.6 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.2 | 7.3 | GO:0051693 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) actin filament capping(GO:0051693) |
| 0.2 | 0.7 | GO:0070178 | D-amino acid catabolic process(GO:0019478) D-serine catabolic process(GO:0036088) D-amino acid metabolic process(GO:0046416) D-serine metabolic process(GO:0070178) |
| 0.2 | 1.3 | GO:0006283 | transcription-coupled nucleotide-excision repair(GO:0006283) |
| 0.2 | 1.4 | GO:1900006 | positive regulation of dendrite development(GO:1900006) |
| 0.2 | 0.7 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.2 | 2.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 3.9 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.2 | 0.7 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.2 | 1.8 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.2 | 4.4 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.2 | 0.5 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.2 | 0.7 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.2 | 1.3 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.2 | 0.9 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.2 | 2.5 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.2 | 0.6 | GO:0061355 | Wnt protein secretion(GO:0061355) |
| 0.2 | 0.6 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.2 | 2.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 1.4 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.2 | 1.1 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 0.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 1.0 | GO:0060386 | synapse assembly involved in innervation(GO:0060386) |
| 0.1 | 1.7 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.1 | 1.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 2.4 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.1 | 1.8 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.1 | 1.7 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.4 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.1 | 4.2 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 13.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 3.7 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.1 | 8.2 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.1 | 3.5 | GO:0043049 | otic placode formation(GO:0043049) ectodermal placode formation(GO:0060788) |
| 0.1 | 0.9 | GO:0045665 | negative regulation of neuron differentiation(GO:0045665) |
| 0.1 | 0.4 | GO:0045905 | positive regulation of protein complex disassembly(GO:0043243) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.5 | GO:0015860 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.1 | 4.5 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.1 | 4.1 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 2.8 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 1.6 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.1 | 0.9 | GO:0006007 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 2.6 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 1.7 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 0.6 | GO:1990402 | cellular response to prostaglandin stimulus(GO:0071379) cellular response to fatty acid(GO:0071398) embryonic liver development(GO:1990402) |
| 0.1 | 0.4 | GO:0032241 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.1 | 1.8 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 0.8 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.1 | 0.4 | GO:0090200 | regulation of release of cytochrome c from mitochondria(GO:0090199) positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.3 | GO:0006404 | RNA import into nucleus(GO:0006404) snRNA import into nucleus(GO:0061015) |
| 0.1 | 1.1 | GO:0098703 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 1.5 | GO:0072160 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) nephron tubule epithelial cell differentiation(GO:0072160) |
| 0.1 | 1.4 | GO:1902807 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.1 | 1.4 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 1.1 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.1 | 0.3 | GO:0072314 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.1 | 1.0 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 0.9 | GO:0032262 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) CTP salvage(GO:0044211) |
| 0.1 | 0.3 | GO:0051881 | regulation of mitochondrial membrane potential(GO:0051881) |
| 0.1 | 1.8 | GO:0032958 | inositol phosphate biosynthetic process(GO:0032958) |
| 0.1 | 0.3 | GO:0042723 | thiamine metabolic process(GO:0006772) thiamine diphosphate metabolic process(GO:0042357) thiamine-containing compound metabolic process(GO:0042723) |
| 0.1 | 0.5 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.1 | 0.4 | GO:0006958 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.1 | 1.7 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.5 | GO:0099640 | dendritic transport(GO:0098935) anterograde dendritic transport(GO:0098937) anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) axo-dendritic protein transport(GO:0099640) |
| 0.1 | 1.4 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.1 | 3.1 | GO:0036303 | lymph vessel morphogenesis(GO:0036303) |
| 0.1 | 0.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 4.9 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 1.7 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.1 | 1.1 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.1 | 1.1 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.1 | 0.8 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.1 | 1.9 | GO:0035725 | sodium ion transmembrane transport(GO:0035725) |
| 0.1 | 0.4 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.1 | 0.4 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 2.4 | GO:0031167 | rRNA methylation(GO:0031167) |
| 0.1 | 2.1 | GO:0043588 | skin development(GO:0043588) |
| 0.1 | 1.1 | GO:0032924 | activin receptor signaling pathway(GO:0032924) |
| 0.1 | 1.3 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.1 | 1.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 3.7 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 8.0 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.1 | 0.8 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 1.2 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.1 | 2.5 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 3.2 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.1 | 3.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.5 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.1 | 1.9 | GO:0072310 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.1 | 1.7 | GO:0097581 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.1 | 0.8 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.4 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.1 | 1.1 | GO:0008217 | regulation of blood pressure(GO:0008217) |
| 0.1 | 2.4 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.1 | 0.9 | GO:0014046 | dopamine secretion(GO:0014046) regulation of dopamine secretion(GO:0014059) dopamine transport(GO:0015872) |
| 0.1 | 1.3 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.1 | 1.0 | GO:0098742 | cell-cell adhesion via plasma-membrane adhesion molecules(GO:0098742) |
| 0.1 | 0.9 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.1 | 0.2 | GO:0003431 | growth plate cartilage chondrocyte development(GO:0003431) chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.1 | 0.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.1 | 1.1 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) regulation of phospholipid metabolic process(GO:1903725) |
| 0.1 | 1.3 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.1 | 1.1 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.3 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 1.8 | GO:0007596 | blood coagulation(GO:0007596) hemostasis(GO:0007599) |
| 0.1 | 1.2 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.1 | 0.5 | GO:0010885 | cholesterol storage(GO:0010878) regulation of cholesterol storage(GO:0010885) |
| 0.1 | 2.0 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.1 | 2.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 2.4 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.1 | 1.6 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.1 | 0.5 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 0.8 | GO:0060402 | cytosolic calcium ion transport(GO:0060401) calcium ion transport into cytosol(GO:0060402) |
| 0.1 | 3.8 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.1 | 2.2 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 0.8 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 0.5 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.1 | 2.0 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.1 | 0.7 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.1 | 0.6 | GO:0009746 | response to carbohydrate(GO:0009743) response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.1 | 1.6 | GO:0045214 | sarcomere organization(GO:0045214) |
| 0.1 | 0.6 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 2.2 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.1 | 0.9 | GO:0060900 | embryonic camera-type eye formation(GO:0060900) |
| 0.1 | 0.2 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.1 | 0.7 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 1.1 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 0.7 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.3 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.3 | GO:0039689 | viral genome replication(GO:0019079) negative stranded viral RNA replication(GO:0039689) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 3.9 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.3 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.7 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.0 | 0.1 | GO:0051043 | regulation of cellular extravasation(GO:0002691) positive regulation of cellular extravasation(GO:0002693) cellular extravasation(GO:0045123) regulation of membrane protein ectodomain proteolysis(GO:0051043) positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.0 | 0.4 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.0 | 0.5 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.2 | GO:1901655 | cellular response to ketone(GO:1901655) |
| 0.0 | 1.1 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.4 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.8 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.6 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 0.4 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.7 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.4 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.2 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.8 | GO:0045103 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 1.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0055062 | cellular anion homeostasis(GO:0030002) cellular monovalent inorganic anion homeostasis(GO:0030320) cellular phosphate ion homeostasis(GO:0030643) phosphate ion homeostasis(GO:0055062) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) trivalent inorganic anion homeostasis(GO:0072506) |
| 0.0 | 0.2 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 0.7 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 1.1 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.2 | GO:0006083 | acetate metabolic process(GO:0006083) acetyl-CoA biosynthetic process from acetate(GO:0019427) |
| 0.0 | 1.5 | GO:0098656 | anion transmembrane transport(GO:0098656) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.6 | GO:0031589 | cell-substrate adhesion(GO:0031589) |
| 0.0 | 0.1 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 1.5 | GO:1901342 | regulation of vasculature development(GO:1901342) |
| 0.0 | 0.3 | GO:0060261 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.7 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 1.3 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.3 | GO:0045010 | actin nucleation(GO:0045010) |
| 0.0 | 0.2 | GO:0048859 | rhombomere boundary formation(GO:0021654) formation of anatomical boundary(GO:0048859) |
| 0.0 | 0.6 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.6 | GO:0048545 | response to steroid hormone(GO:0048545) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.2 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.1 | GO:0043620 | regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) regulation of DNA-templated transcription in response to stress(GO:0043620) |
| 0.0 | 0.6 | GO:0035272 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 0.3 | GO:0045740 | positive regulation of DNA replication(GO:0045740) |
| 0.0 | 0.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.8 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 0.2 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.2 | GO:0035308 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.6 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 1.8 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.8 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.0 | GO:0046546 | development of primary male sexual characteristics(GO:0046546) male sex differentiation(GO:0046661) |
| 0.0 | 0.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.2 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.6 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.2 | GO:0016358 | dendrite development(GO:0016358) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 8.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.8 | 4.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.8 | 5.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.7 | 16.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.6 | 3.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.5 | 2.2 | GO:0098833 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) extrinsic component of presynaptic membrane(GO:0098888) extrinsic component of presynaptic endocytic zone(GO:0098894) |
| 0.5 | 1.5 | GO:0031362 | intrinsic component of external side of plasma membrane(GO:0031233) anchored component of external side of plasma membrane(GO:0031362) |
| 0.5 | 3.4 | GO:0031838 | hemoglobin complex(GO:0005833) haptoglobin-hemoglobin complex(GO:0031838) |
| 0.5 | 2.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.4 | 1.8 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.4 | 3.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.4 | 1.2 | GO:0035101 | FACT complex(GO:0035101) |
| 0.4 | 4.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.4 | 1.5 | GO:0016586 | RSC complex(GO:0016586) |
| 0.4 | 1.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.4 | 2.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.3 | 2.1 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 1.4 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.3 | 1.4 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.3 | 5.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 2.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.2 | 1.2 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 2.8 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.2 | 2.3 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.2 | 0.9 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.2 | 0.7 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 1.8 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 39.3 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.2 | 1.2 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.2 | 2.0 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.2 | 3.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 1.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 2.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.2 | 2.5 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.2 | 5.6 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.2 | 3.6 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.2 | 1.8 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.2 | 0.7 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.2 | 0.5 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.2 | 1.8 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.2 | 0.8 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.2 | 0.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.2 | 0.8 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 2.1 | GO:0045178 | basal part of cell(GO:0045178) basal cortex(GO:0045180) |
| 0.1 | 1.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 5.6 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.1 | 2.8 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.1 | 3.1 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 1.8 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.1 | 0.9 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 2.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 6.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 5.7 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 4.7 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 1.3 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.1 | 2.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 7.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 3.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 1.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.3 | GO:1990879 | CST complex(GO:1990879) |
| 0.1 | 5.5 | GO:0031674 | I band(GO:0031674) |
| 0.1 | 2.9 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.9 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 4.3 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.1 | 1.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.9 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 11.3 | GO:0043005 | neuron projection(GO:0043005) |
| 0.1 | 2.6 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 1.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.7 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 5.3 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.1 | 0.2 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 1.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 2.0 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.9 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 3.2 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 6.9 | GO:0030425 | dendrite(GO:0030425) |
| 0.1 | 1.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 38.5 | GO:0031226 | intrinsic component of plasma membrane(GO:0031226) |
| 0.1 | 0.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 1.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.1 | 1.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 2.3 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 7.7 | GO:0009986 | cell surface(GO:0009986) |
| 0.1 | 1.9 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.1 | 2.4 | GO:0030136 | clathrin-coated vesicle(GO:0030136) |
| 0.1 | 2.3 | GO:0030141 | secretory granule(GO:0030141) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0033181 | plasma membrane proton-transporting V-type ATPase complex(GO:0033181) |
| 0.0 | 0.7 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 2.3 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 0.4 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 1.4 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 0.2 | GO:0044453 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 1.8 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.1 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.0 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 1.7 | GO:0031410 | cytoplasmic vesicle(GO:0031410) |
| 0.0 | 0.2 | GO:0001725 | stress fiber(GO:0001725) actomyosin(GO:0042641) contractile actin filament bundle(GO:0097517) |
| 0.0 | 7.9 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 3.0 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.1 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 5.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.6 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.9 | 6.4 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.9 | 7.1 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.8 | 4.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.8 | 3.0 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.7 | 3.7 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.7 | 2.8 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.7 | 2.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.7 | 4.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.6 | 1.8 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.6 | 2.4 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.6 | 3.0 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.6 | 4.1 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) lipoprotein particle receptor binding(GO:0070325) |
| 0.6 | 3.4 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.6 | 1.7 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.5 | 3.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.5 | 4.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.5 | 1.5 | GO:0008465 | glycerate dehydrogenase activity(GO:0008465) |
| 0.5 | 1.5 | GO:0102344 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.5 | 7.8 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.5 | 3.4 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.5 | 5.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.5 | 6.4 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.5 | 2.3 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.4 | 4.4 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.4 | 4.8 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.4 | 3.0 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.4 | 2.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) |
| 0.4 | 11.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.4 | 3.6 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.4 | 1.5 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.4 | 1.1 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.4 | 2.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.4 | 2.9 | GO:0005522 | profilin binding(GO:0005522) |
| 0.4 | 1.4 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.4 | 4.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.3 | 1.4 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.3 | 3.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.3 | 1.4 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.3 | 1.3 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.3 | 1.9 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.3 | 2.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.3 | 2.5 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.3 | 1.8 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.3 | 1.2 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.3 | 2.4 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.3 | 6.6 | GO:0042805 | actinin binding(GO:0042805) |
| 0.3 | 13.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.3 | 0.9 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.3 | 1.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.3 | 1.1 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.3 | 1.3 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.3 | 3.7 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.3 | 1.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 3.1 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.3 | 1.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.2 | 3.7 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 2.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.2 | 0.7 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.2 | 14.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 1.0 | GO:0099528 | G-protein coupled neurotransmitter receptor activity(GO:0099528) |
| 0.2 | 6.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.2 | 1.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.2 | 1.2 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 0.7 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.2 | 1.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.1 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.2 | 1.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.2 | 2.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 1.3 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.2 | 1.9 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.2 | 0.8 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.2 | 1.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.2 | 0.6 | GO:0039706 | co-receptor binding(GO:0039706) receptor antagonist activity(GO:0048019) |
| 0.2 | 1.9 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.2 | 4.9 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.2 | 2.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 1.0 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.2 | 1.8 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.2 | 0.6 | GO:0004953 | icosanoid receptor activity(GO:0004953) |
| 0.2 | 1.4 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 1.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.2 | 0.8 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.2 | 2.3 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 1.9 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 2.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.2 | 0.7 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.2 | 0.7 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.2 | 0.7 | GO:0098918 | structural constituent of presynaptic active zone(GO:0098882) structural constituent of synapse(GO:0098918) |
| 0.2 | 0.7 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.2 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.2 | 3.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 1.3 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.2 | 0.8 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.2 | 0.7 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.2 | 0.6 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.2 | 1.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.2 | 3.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.2 | 2.3 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 0.9 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.1 | 2.2 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.1 | 1.8 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) |
| 0.1 | 2.7 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 10.8 | GO:0098531 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.1 | 2.6 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 3.1 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.1 | 0.6 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 2.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 0.4 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.9 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.5 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.1 | 3.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 0.6 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.1 | 2.4 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.9 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 10.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.7 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.1 | 0.2 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.1 | 1.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 2.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.8 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 1.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.1 | 4.2 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.9 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.1 | 0.5 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.1 | 1.3 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.1 | 1.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 1.0 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
| 0.1 | 0.5 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 1.4 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.7 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.1 | 3.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.5 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 4.8 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.1 | 1.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.1 | 1.1 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.5 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 1.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 1.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.3 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.1 | 1.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.8 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.1 | 1.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 1.1 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 0.9 | GO:0030545 | receptor regulator activity(GO:0030545) |
| 0.1 | 0.4 | GO:0046922 | peptide-O-fucosyltransferase activity(GO:0046922) |
| 0.1 | 0.6 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.2 | GO:0044620 | ACP phosphopantetheine attachment site binding involved in fatty acid biosynthetic process(GO:0000036) ACP phosphopantetheine attachment site binding(GO:0044620) prosthetic group binding(GO:0051192) |
| 0.1 | 1.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.1 | 1.6 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 1.7 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.1 | 2.3 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 2.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 1.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.2 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.1 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.1 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 2.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 27.1 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 2.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 1.2 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 1.7 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 0.2 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.8 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.8 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 10.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 1.3 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.9 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.5 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 1.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 48.3 | GO:0001159 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 1.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0019870 | chloride channel regulator activity(GO:0017081) chloride channel inhibitor activity(GO:0019869) potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 1.9 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 12.4 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.3 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 1.1 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.4 | GO:0022841 | leak channel activity(GO:0022840) potassium ion leak channel activity(GO:0022841) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.2 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.7 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.5 | GO:0019201 | nucleotide kinase activity(GO:0019201) |
| 0.0 | 1.1 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 1.0 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.6 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 1.1 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.0 | 0.2 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.6 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.8 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.5 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 0.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.7 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.2 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 0.7 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.0 | 1.2 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 4.5 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 7.9 | GO:0001071 | nucleic acid binding transcription factor activity(GO:0001071) transcription factor activity, sequence-specific DNA binding(GO:0003700) |
| 0.0 | 0.2 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.3 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 2.4 | GO:0038023 | signaling receptor activity(GO:0038023) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.3 | 6.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.3 | 0.8 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.2 | 6.4 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.2 | 1.9 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.2 | 1.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.2 | 2.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 1.0 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.2 | 2.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 4.2 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 4.9 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 1.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 4.1 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 1.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 3.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.1 | 0.2 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.1 | 2.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 1.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.1 | 2.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 5.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.1 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 2.7 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.3 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 3.3 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 1.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 2.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.5 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.6 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.6 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.2 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.4 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.2 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID ATR PATHWAY | ATR signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 5.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 1.1 | 6.7 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.8 | 1.6 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.5 | 3.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.5 | 3.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.5 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.3 | 0.7 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.3 | 2.4 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.3 | 5.0 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.3 | 1.0 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 1.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.3 | 3.3 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.3 | 1.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.3 | 3.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.2 | 3.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 0.9 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.2 | 2.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.2 | 1.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 1.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 1.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.2 | 0.3 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.2 | 1.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.2 | 2.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.2 | 1.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.2 | 1.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 0.8 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.1 | 6.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 1.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 2.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 1.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.7 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.5 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 1.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.1 | 0.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 1.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 0.2 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.1 | 0.2 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 1.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.1 | 0.6 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 1.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.1 | 0.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 2.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 0.6 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 0.7 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 3.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.9 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.6 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 1.6 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 1.1 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.4 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 1.1 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.8 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 1.0 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.3 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 1.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.9 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.8 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.3 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.1 | REACTOME HOST INTERACTIONS OF HIV FACTORS | Genes involved in Host Interactions of HIV factors |
| 0.0 | 0.7 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.2 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.5 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |