Project
DANIO-CODE
Navigation
Downloads
Results for sox32
Z-value: 0.83
Transcription factors associated with sox32
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox32
|
ENSDARG00000100591 | SRY-box transcription factor 32 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox32 | dr10_dc_chr7_-_58524285_58524309 | -0.13 | 6.3e-01 | Click! |
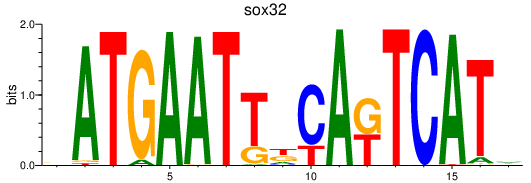
Activity profile of sox32 motif
Sorted Z-values of sox32 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of sox32
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_11113032 | 3.62 |
ENSDART00000027598
|
tpm3
|
tropomyosin 3 |
| chr15_-_23711689 | 3.27 |
ENSDART00000128644
|
ckmb
|
creatine kinase, muscle b |
| chr23_-_26150495 | 2.87 |
ENSDART00000126299
|
gdi1
|
GDP dissociation inhibitor 1 |
| chr2_+_33057931 | 2.61 |
|
|
|
| chr21_-_41286846 | 2.45 |
ENSDART00000167339
|
msx2b
|
muscle segment homeobox 2b |
| chr12_+_29994735 | 2.40 |
ENSDART00000042572
ENSDART00000153025 |
ablim1b
|
actin binding LIM protein 1b |
| chr13_-_25067585 | 2.13 |
ENSDART00000159585
|
adka
|
adenosine kinase a |
| chr1_-_49004615 | 1.94 |
ENSDART00000134399
|
slkb
|
STE20-like kinase b |
| chr17_+_27417635 | 1.91 |
ENSDART00000052446
|
vgll2b
|
vestigial-like family member 2b |
| chr9_+_11616740 | 1.89 |
ENSDART00000146585
|
si:dkey-69o16.5
|
si:dkey-69o16.5 |
| chr15_-_35394535 | 1.77 |
ENSDART00000144153
|
mff
|
mitochondrial fission factor |
| chr10_+_4962403 | 1.76 |
ENSDART00000134679
|
CU074419.2
|
ENSDARG00000093688 |
| chr25_-_17483202 | 1.68 |
ENSDART00000073684
|
mmp15a
|
matrix metallopeptidase 15a |
| KN149968v1_+_15749 | 1.50 |
ENSDART00000168977
|
ENSDARG00000102503
|
ENSDARG00000102503 |
| chr24_-_36938495 | 1.50 |
ENSDART00000112694
|
fam171a2b
|
family with sequence similarity 171, member A2b |
| chr25_-_28054271 | 1.45 |
|
|
|
| chr25_-_22541525 | 1.40 |
ENSDART00000073583
|
islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr11_+_10557530 | 1.27 |
ENSDART00000132365
|
b3gnt5a
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5a |
| chr19_-_42892975 | 1.24 |
ENSDART00000131715
|
psmb4
|
proteasome subunit beta 4 |
| chr14_+_6239755 | 1.16 |
ENSDART00000097214
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr24_+_35899507 | 1.16 |
ENSDART00000122408
|
si:dkeyp-7a3.1
|
si:dkeyp-7a3.1 |
| chr2_-_42784609 | 1.15 |
|
|
|
| chr10_+_31362144 | 1.13 |
|
|
|
| chr8_+_13465718 | 1.09 |
ENSDART00000034740
|
fut9d
|
fucosyltransferase 9d |
| chr5_-_67233396 | 1.09 |
ENSDART00000051833
ENSDART00000124890 |
gsx1
|
GS homeobox 1 |
| chr19_-_19883594 | 1.01 |
|
|
|
| chr19_+_5562107 | 0.99 |
ENSDART00000082080
|
jupb
|
junction plakoglobin b |
| chr10_-_7954888 | 0.98 |
ENSDART00000139661
|
slc35e4
|
solute carrier family 35, member E4 |
| chr19_-_10941519 | 0.97 |
ENSDART00000169118
ENSDART00000160599 |
CU855694.1
|
ENSDARG00000098513 |
| chr18_-_40493952 | 0.94 |
|
|
|
| chr12_+_26530620 | 0.81 |
ENSDART00000046959
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr7_-_18448121 | 0.75 |
ENSDART00000112359
|
si:ch211-119e14.9
|
si:ch211-119e14.9 |
| chr11_+_44108192 | 0.72 |
ENSDART00000165219
ENSDART00000160678 |
srsf7b
|
serine/arginine-rich splicing factor 7b |
| chr2_-_25340798 | 0.70 |
ENSDART00000169711
ENSDART00000144795 ENSDART00000111507 |
slc35g2a
|
solute carrier family 35, member G2a |
| chr25_-_22541628 | 0.68 |
ENSDART00000163001
|
islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr12_+_29994880 | 0.67 |
ENSDART00000042572
ENSDART00000153025 |
ablim1b
|
actin binding LIM protein 1b |
| KN150630v1_+_44324 | 0.66 |
|
|
|
| chr15_-_37949371 | 0.65 |
ENSDART00000031418
|
hsc70
|
heat shock cognate 70 |
| chr6_+_44891278 | 0.65 |
|
|
|
| chr8_+_16690670 | 0.64 |
ENSDART00000144621
|
smim15
|
small integral membrane protein 15 |
| chr2_-_27730279 | 0.57 |
ENSDART00000147100
|
toe1
|
target of EGR1, member 1 (nuclear) |
| chr3_-_53467621 | 0.55 |
ENSDART00000032788
|
rdh8a
|
retinol dehydrogenase 8a |
| chr21_-_19293600 | 0.54 |
ENSDART00000101462
|
mrps18c
|
mitochondrial ribosomal protein S18C |
| chr20_-_4723724 | 0.51 |
ENSDART00000147071
|
ak7a
|
adenylate kinase 7a |
| chr22_+_699548 | 0.47 |
ENSDART00000017305
|
znf76
|
zinc finger protein 76 |
| chr19_-_29849858 | 0.47 |
ENSDART00000052108
ENSDART00000171908 |
fndc5b
|
fibronectin type III domain containing 5b |
| chr2_+_17129963 | 0.44 |
ENSDART00000145778
|
eif4g1a
|
eukaryotic translation initiation factor 4 gamma, 1a |
| chr19_-_29849805 | 0.42 |
ENSDART00000114442
|
fndc5b
|
fibronectin type III domain containing 5b |
| chr9_+_33049445 | 0.40 |
ENSDART00000168992
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr20_+_571020 | 0.40 |
ENSDART00000152149
|
si:dkey-121j17.6
|
si:dkey-121j17.6 |
| chr9_-_23222916 | 0.39 |
ENSDART00000022392
ENSDART00000122036 |
rnd3b
|
Rho family GTPase 3b |
| chr9_+_11616570 | 0.38 |
ENSDART00000146585
|
si:dkey-69o16.5
|
si:dkey-69o16.5 |
| chr4_+_27111238 | 0.37 |
ENSDART00000141908
|
alg12
|
ALG12, alpha-1,6-mannosyltransferase |
| chr2_-_42784707 | 0.37 |
|
|
|
| chr9_+_33049549 | 0.34 |
ENSDART00000121751
|
si:dkey-145p14.5
|
si:dkey-145p14.5 |
| chr7_+_17923953 | 0.34 |
ENSDART00000109171
|
rce1a
|
Ras converting CAAX endopeptidase 1a |
| chr5_-_1296190 | 0.34 |
|
|
|
| chr16_+_40074850 | 0.28 |
ENSDART00000110100
|
hint3
|
histidine triad nucleotide binding protein 3 |
| chr10_+_15106739 | 0.22 |
ENSDART00000128967
|
parm1
|
prostate androgen-regulated mucin-like protein 1 |
| chr22_-_12837665 | 0.20 |
ENSDART00000145156
ENSDART00000137280 |
glsa
|
glutaminase a |
| chr21_-_32244805 | 0.15 |
ENSDART00000178765
|
BX927388.2
|
ENSDARG00000107266 |
| chr1_+_16983711 | 0.09 |
ENSDART00000030665
|
slc25a4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 |
| chr7_+_41141295 | 0.07 |
ENSDART00000083967
|
chpf2
|
chondroitin polymerizing factor 2 |
| chr22_+_5652765 | 0.06 |
ENSDART00000131308
|
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr15_-_21756778 | 0.06 |
ENSDART00000039865
|
sdhdb
|
succinate dehydrogenase complex, subunit D, integral membrane protein b |
| chr8_-_16614882 | 0.05 |
ENSDART00000135319
|
osbpl9
|
oxysterol binding protein-like 9 |
| chr10_+_31362087 | 0.04 |
|
|
|
| chr7_-_52113622 | 0.03 |
ENSDART00000174186
|
wdr93
|
WD repeat domain 93 |
| chr19_+_10941495 | 0.00 |
ENSDART00000169962
|
si:ch73-347e22.4
|
si:ch73-347e22.4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.4 | 3.3 | GO:0006603 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.2 | 1.1 | GO:0036065 | fucosylation(GO:0036065) |
| 0.1 | 1.7 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.1 | 3.1 | GO:0097581 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.1 | 3.6 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.1 | 0.5 | GO:0008210 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.1 | 0.7 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 1.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 1.2 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.4 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.2 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 2.1 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.2 | GO:0043650 | dicarboxylic acid biosynthetic process(GO:0043650) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.6 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0015860 | intracellular nucleoside transport(GO:0015859) purine nucleoside transmembrane transport(GO:0015860) purine nucleotide transport(GO:0015865) ATP transport(GO:0015867) purine ribonucleotide transport(GO:0015868) adenine nucleotide transport(GO:0051503) purine-containing compound transmembrane transport(GO:0072530) mitochondrial ATP transmembrane transport(GO:1990544) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.5 | GO:0009142 | nucleoside triphosphate biosynthetic process(GO:0009142) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.1 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.1 | 1.2 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 3.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 1.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.9 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.5 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.4 | 3.3 | GO:0004111 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.4 | 1.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.2 | 2.9 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.1 | 0.5 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 0.5 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 1.2 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.2 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.7 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 1.3 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.4 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 3.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 6.7 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.1 | GO:0005471 | ATP:ADP antiporter activity(GO:0005471) |
| 0.0 | 2.1 | GO:0017171 | serine-type peptidase activity(GO:0008236) serine hydrolase activity(GO:0017171) |
| 0.0 | 0.9 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.6 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 1.0 | GO:0015297 | antiporter activity(GO:0015297) |
| 0.0 | 0.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 2.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.2 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |