Project
DANIO-CODE
Navigation
Downloads
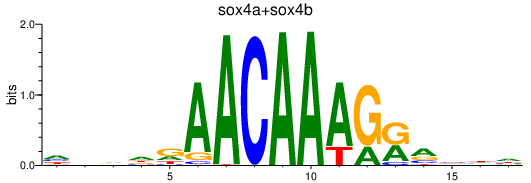
Results for sox4a+sox4b
Z-value: 0.74
Transcription factors associated with sox4a+sox4b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox4a
|
ENSDARG00000004588 | SRY-box transcription factor 4a |
|
sox4b
|
ENSDARG00000098834 | SRY-box transcription factor 4b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox4a | dr10_dc_chr19_-_29205158_29205231 | -0.65 | 6.1e-03 | Click! |
| sox4b | dr10_dc_chr16_+_68014_68039 | -0.39 | 1.4e-01 | Click! |
Activity profile of sox4a+sox4b motif
Sorted Z-values of sox4a+sox4b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of sox4a+sox4b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_43471879 | 1.25 |
ENSDART00000166324
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr14_-_32755189 | 1.21 |
ENSDART00000074720
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr14_+_14850200 | 1.18 |
ENSDART00000167966
|
zgc:158852
|
zgc:158852 |
| chr12_+_30673985 | 1.17 |
ENSDART00000160422
|
aldh18a1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr8_+_29626848 | 1.14 |
ENSDART00000139029
ENSDART00000091409 |
smarcad1a
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 a |
| chr16_+_35582277 | 1.02 |
ENSDART00000167001
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr16_+_35582648 | 1.01 |
ENSDART00000171675
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr23_-_36349563 | 1.00 |
|
|
|
| chr5_+_3567992 | 0.99 |
ENSDART00000129329
|
rpain
|
RPA interacting protein |
| chr22_-_6995611 | 0.99 |
ENSDART00000003422
|
smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr3_-_27515161 | 0.95 |
ENSDART00000151027
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr3_-_15960491 | 0.92 |
ENSDART00000064838
|
lasp1
|
LIM and SH3 protein 1 |
| chr14_+_38509671 | 0.88 |
|
|
|
| chr1_+_43471798 | 0.87 |
ENSDART00000166324
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr8_-_44617324 | 0.86 |
ENSDART00000063396
|
bag4
|
BCL2-associated athanogene 4 |
| chr7_-_64745132 | 0.86 |
ENSDART00000112166
|
fam60al
|
family with sequence similarity 60, member A, like |
| chr23_-_34017934 | 0.84 |
ENSDART00000133223
|
ENSDARG00000070393
|
ENSDARG00000070393 |
| chr16_-_47446494 | 0.83 |
ENSDART00000032188
|
si:dkey-256h2.1
|
si:dkey-256h2.1 |
| chr7_-_69883542 | 0.82 |
|
|
|
| chr7_-_24724911 | 0.82 |
ENSDART00000173791
|
rcor2
|
REST corepressor 2 |
| chr23_-_35384257 | 0.80 |
ENSDART00000113643
|
fbxo25
|
F-box protein 25 |
| chr17_+_7377230 | 0.79 |
ENSDART00000157123
|
shprh
|
SNF2 histone linker PHD RING helicase |
| chr24_+_19270877 | 0.78 |
|
|
|
| chr5_-_22099244 | 0.77 |
ENSDART00000161298
|
nono
|
non-POU domain containing, octamer-binding |
| chr14_+_38510523 | 0.77 |
|
|
|
| chr5_+_22541236 | 0.77 |
ENSDART00000171719
|
atrxl
|
alpha thalassemia/mental retardation syndrome X-linked, like |
| chr14_-_32755454 | 0.76 |
ENSDART00000074720
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr21_-_37341980 | 0.75 |
ENSDART00000076483
|
zgc:77151
|
zgc:77151 |
| chr8_-_44617367 | 0.73 |
ENSDART00000063396
|
bag4
|
BCL2-associated athanogene 4 |
| chr1_-_22617455 | 0.73 |
ENSDART00000137567
|
smim14
|
small integral membrane protein 14 |
| chr7_-_31346802 | 0.73 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr4_-_25921887 | 0.72 |
ENSDART00000175487
|
fgd6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr3_+_60299938 | 0.70 |
ENSDART00000053482
|
tsen54
|
TSEN54 tRNA splicing endonuclease subunit |
| chr5_+_44246311 | 0.69 |
ENSDART00000145299
ENSDART00000136521 |
kank1a
|
KN motif and ankyrin repeat domains 1a |
| chr7_-_69883581 | 0.69 |
|
|
|
| chr16_-_33151404 | 0.68 |
ENSDART00000145055
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr7_-_31346844 | 0.67 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr1_+_18390488 | 0.67 |
ENSDART00000002886
ENSDART00000158745 |
exosc9
|
exosome component 9 |
| chr4_-_25921780 | 0.66 |
ENSDART00000175487
|
fgd6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr2_-_19585211 | 0.65 |
ENSDART00000169384
|
cdc20
|
cell division cycle 20 homolog |
| chr16_+_35582411 | 0.64 |
ENSDART00000171675
|
cited4b
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4b |
| chr25_+_15176959 | 0.62 |
ENSDART00000045659
|
tcp11l1
|
t-complex 11, testis-specific-like 1 |
| chr5_-_66070385 | 0.62 |
ENSDART00000032909
|
kat5b
|
K(lysine) acetyltransferase 5b |
| chr16_-_55211053 | 0.62 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr19_-_18689437 | 0.61 |
ENSDART00000016135
|
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr24_-_21828216 | 0.59 |
ENSDART00000030592
|
acot9.1
|
acyl-CoA thioesterase 9, tandem duplicate 1 |
| chr5_-_18393623 | 0.59 |
ENSDART00000010101
ENSDART00000099434 |
aacs
|
acetoacetyl-CoA synthetase |
| chr7_-_73628151 | 0.59 |
ENSDART00000123136
|
FP236812.4
|
Histone H2B 1/2 |
| chr23_-_16755868 | 0.59 |
ENSDART00000020810
|
sdcbp2
|
syndecan binding protein (syntenin) 2 |
| chr20_+_6543305 | 0.58 |
ENSDART00000135005
ENSDART00000166356 |
si:ch211-191a24.4
|
si:ch211-191a24.4 |
| chr20_-_3301951 | 0.58 |
ENSDART00000123096
|
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr1_+_43471906 | 0.58 |
ENSDART00000166324
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr1_+_43472177 | 0.57 |
ENSDART00000170512
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr7_+_10320931 | 0.56 |
ENSDART00000173125
|
zfand6
|
zinc finger, AN1-type domain 6 |
| chr1_+_43471683 | 0.56 |
ENSDART00000166324
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr6_-_6818607 | 0.56 |
ENSDART00000151822
|
tuba8l4
|
tubulin, alpha 8 like 4 |
| chr19_+_7717962 | 0.54 |
ENSDART00000112404
|
cgnb
|
cingulin b |
| chr1_+_43471754 | 0.53 |
ENSDART00000166324
|
ctnnd1
|
catenin (cadherin-associated protein), delta 1 |
| chr8_+_29626894 | 0.53 |
ENSDART00000139029
ENSDART00000091409 |
smarcad1a
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 a |
| chr4_-_25921852 | 0.52 |
ENSDART00000175487
|
fgd6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr5_-_27549500 | 0.52 |
ENSDART00000051515
|
zgc:110329
|
zgc:110329 |
| chr13_-_43503453 | 0.52 |
ENSDART00000127930
ENSDART00000084474 |
fam160b1
|
family with sequence similarity 160, member B1 |
| chr3_+_30187036 | 0.52 |
ENSDART00000151006
|
CR936968.1
|
ENSDARG00000096295 |
| chr6_-_9046536 | 0.51 |
ENSDART00000159506
|
ccdc14
|
coiled-coil domain containing 14 |
| chr23_-_3815871 | 0.50 |
ENSDART00000137826
|
hmga1a
|
high mobility group AT-hook 1a |
| chr5_-_22466262 | 0.50 |
ENSDART00000172549
|
si:dkey-237j10.2
|
si:dkey-237j10.2 |
| chr12_-_22279843 | 0.50 |
ENSDART00000153194
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr20_-_3301898 | 0.49 |
ENSDART00000123096
|
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr6_-_8008902 | 0.49 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr4_-_15005050 | 0.49 |
ENSDART00000067048
|
klhdc10
|
kelch domain containing 10 |
| chr5_+_20319483 | 0.49 |
ENSDART00000142894
|
limk2
|
LIM domain kinase 2 |
| chr16_+_42567707 | 0.48 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr24_+_26223767 | 0.48 |
|
|
|
| chr12_-_22279890 | 0.48 |
ENSDART00000153194
|
si:dkey-38p12.3
|
si:dkey-38p12.3 |
| chr16_+_26858575 | 0.47 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr23_-_10810190 | 0.47 |
ENSDART00000140745
|
eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr18_+_18416922 | 0.46 |
ENSDART00000080174
|
n4bp1
|
nedd4 binding protein 1 |
| chr25_+_15177176 | 0.46 |
ENSDART00000045659
|
tcp11l1
|
t-complex 11, testis-specific-like 1 |
| chr8_-_44617200 | 0.44 |
ENSDART00000063396
|
bag4
|
BCL2-associated athanogene 4 |
| chr14_+_38510427 | 0.44 |
|
|
|
| chr1_+_30840656 | 0.44 |
ENSDART00000075286
|
slc2a15b
|
solute carrier family 2 (facilitated glucose transporter), member 15b |
| chr22_-_7976467 | 0.44 |
ENSDART00000162028
|
sc:d217
|
sc:d217 |
| chr1_-_46260853 | 0.43 |
|
|
|
| chr2_-_24413154 | 0.43 |
ENSDART00000145526
|
slc12a7a
|
solute carrier family 12 (potassium/chloride transporter), member 7a |
| chr12_+_10078043 | 0.43 |
ENSDART00000152369
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr3_+_18657831 | 0.42 |
ENSDART00000055757
|
tnpo2
|
transportin 2 (importin 3, karyopherin beta 2b) |
| chr5_-_18393705 | 0.41 |
ENSDART00000010101
ENSDART00000099434 |
aacs
|
acetoacetyl-CoA synthetase |
| chr2_+_43355846 | 0.41 |
ENSDART00000056402
|
pard3ab
|
par-3 family cell polarity regulator alpha, b |
| chr7_-_31669675 | 0.40 |
ENSDART00000131009
|
bdnf
|
brain-derived neurotrophic factor |
| chr3_-_52406525 | 0.40 |
ENSDART00000138018
|
zgc:113210
|
zgc:113210 |
| chr7_-_31346991 | 0.39 |
ENSDART00000111388
|
igdcc3
|
immunoglobulin superfamily, DCC subclass, member 3 |
| chr13_+_15802079 | 0.39 |
ENSDART00000131390
|
fignl1
|
fidgetin-like 1 |
| chr2_+_38019437 | 0.39 |
ENSDART00000139564
|
dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr14_+_14536088 | 0.38 |
ENSDART00000158291
|
slbp
|
stem-loop binding protein |
| chr14_+_6656429 | 0.38 |
ENSDART00000150050
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr9_+_27518890 | 0.38 |
ENSDART00000111039
|
GTPBP8
|
GTP binding protein 8 (putative) |
| chr14_-_32755370 | 0.37 |
ENSDART00000074720
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr13_+_762105 | 0.37 |
|
|
|
| chr19_-_31212648 | 0.36 |
ENSDART00000125893
ENSDART00000145581 |
trit1
|
tRNA isopentenyltransferase 1 |
| chr17_-_49994799 | 0.36 |
|
|
|
| chr1_+_30840599 | 0.36 |
ENSDART00000075286
|
slc2a15b
|
solute carrier family 2 (facilitated glucose transporter), member 15b |
| chr8_+_29626728 | 0.36 |
ENSDART00000139029
ENSDART00000091409 |
smarcad1a
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 a |
| chr23_-_36350288 | 0.36 |
ENSDART00000110478
|
zgc:174906
|
zgc:174906 |
| chr5_-_7591547 | 0.36 |
ENSDART00000158447
|
nipbla
|
nipped-B homolog a (Drosophila) |
| chr7_+_10320888 | 0.36 |
ENSDART00000173125
|
zfand6
|
zinc finger, AN1-type domain 6 |
| chr1_-_17073539 | 0.35 |
|
|
|
| chr8_-_19437095 | 0.35 |
ENSDART00000162010
|
zgc:92140
|
zgc:92140 |
| chr22_-_6995444 | 0.35 |
ENSDART00000003422
|
smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr21_-_37342215 | 0.35 |
ENSDART00000076483
|
zgc:77151
|
zgc:77151 |
| chr18_+_18417187 | 0.35 |
ENSDART00000080174
|
n4bp1
|
nedd4 binding protein 1 |
| chr6_-_8009143 | 0.34 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr12_+_33818555 | 0.34 |
ENSDART00000085888
|
trim8b
|
tripartite motif containing 8b |
| chr16_+_12726556 | 0.34 |
|
|
|
| chr7_-_73628090 | 0.33 |
ENSDART00000123136
|
FP236812.4
|
Histone H2B 1/2 |
| chr17_+_43916865 | 0.33 |
ENSDART00000049503
|
peli2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr7_-_51351777 | 0.33 |
ENSDART00000174111
|
nhsl2
|
NHS-like 2 |
| chr5_+_30880165 | 0.32 |
ENSDART00000098197
|
ENSDARG00000035471
|
ENSDARG00000035471 |
| chr25_+_3381632 | 0.32 |
ENSDART00000157777
|
zgc:153293
|
zgc:153293 |
| chr12_+_19199002 | 0.32 |
ENSDART00000066389
|
tmem184ba
|
transmembrane protein 184ba |
| chr23_-_34017830 | 0.32 |
ENSDART00000133223
|
ENSDARG00000070393
|
ENSDARG00000070393 |
| chr23_-_34018069 | 0.31 |
ENSDART00000133223
|
ENSDARG00000070393
|
ENSDARG00000070393 |
| chr1_-_54089140 | 0.31 |
|
|
|
| chr7_-_73628049 | 0.30 |
ENSDART00000123136
|
FP236812.4
|
Histone H2B 1/2 |
| chr23_-_31883578 | 0.29 |
ENSDART00000139076
|
hbs1l
|
HBS1-like translational GTPase |
| chr5_+_20319434 | 0.29 |
ENSDART00000093185
|
limk2
|
LIM domain kinase 2 |
| chr22_+_38322821 | 0.29 |
ENSDART00000104498
|
rcor3
|
REST corepressor 3 |
| chr23_-_10810264 | 0.29 |
ENSDART00000013768
|
eif4e3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr8_-_37072691 | 0.28 |
ENSDART00000004041
|
zgc:162200
|
zgc:162200 |
| chr11_-_2226711 | 0.28 |
|
|
|
| chr16_+_51317682 | 0.28 |
ENSDART00000157736
|
hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr19_+_7505348 | 0.28 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr14_+_6655894 | 0.28 |
ENSDART00000148394
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr20_-_8847351 | 0.28 |
ENSDART00000141375
|
CR388209.1
|
ENSDARG00000091901 |
| chr25_-_413809 | 0.27 |
|
|
|
| chr5_-_12586284 | 0.26 |
ENSDART00000051664
|
ypel1
|
yippee-like 1 |
| chr13_-_5128884 | 0.26 |
ENSDART00000110610
|
si:dkey-78p8.1
|
si:dkey-78p8.1 |
| chr14_+_6655541 | 0.26 |
ENSDART00000148447
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr17_+_21797517 | 0.25 |
ENSDART00000079011
|
ikzf5
|
IKAROS family zinc finger 5 |
| chr21_-_40759658 | 0.25 |
ENSDART00000075808
|
apbb3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr22_-_7976568 | 0.25 |
ENSDART00000162028
|
sc:d217
|
sc:d217 |
| chr24_-_26224123 | 0.25 |
ENSDART00000080039
|
lrrc31
|
leucine rich repeat containing 31 |
| chr17_+_30529610 | 0.25 |
ENSDART00000154385
|
nhsl1a
|
NHS-like 1a |
| chr19_+_40439831 | 0.24 |
ENSDART00000123647
|
sfpq
|
splicing factor proline/glutamine-rich |
| chr14_-_32755624 | 0.24 |
ENSDART00000074720
|
dlg3
|
discs, large homolog 3 (Drosophila) |
| chr5_-_37281096 | 0.24 |
ENSDART00000084819
|
arhgap35b
|
Rho GTPase activating protein 35b |
| chr11_+_26366532 | 0.24 |
ENSDART00000159505
|
dynlrb1
|
dynein, light chain, roadblock-type 1 |
| chr17_-_7377094 | 0.24 |
|
|
|
| chr23_-_3816422 | 0.24 |
ENSDART00000132205
|
hmga1a
|
high mobility group AT-hook 1a |
| chr12_-_19998765 | 0.24 |
ENSDART00000112768
|
ubald1a
|
UBA-like domain containing 1a |
| chr23_-_17502717 | 0.24 |
|
|
|
| chr11_+_44962456 | 0.23 |
ENSDART00000172792
|
si:ch73-100l22.3
|
si:ch73-100l22.3 |
| chr16_+_9835124 | 0.23 |
ENSDART00000020859
|
pip5k1ab
|
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha, b |
| chr25_-_34619603 | 0.23 |
ENSDART00000114767
|
FQ312024.1
|
ENSDARG00000076129 |
| chr15_+_12497915 | 0.22 |
ENSDART00000168997
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr12_+_33818414 | 0.22 |
ENSDART00000085888
|
trim8b
|
tripartite motif containing 8b |
| chr20_+_13885812 | 0.21 |
ENSDART00000142999
|
lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr17_-_45395144 | 0.21 |
ENSDART00000132969
|
znf106a
|
zinc finger protein 106a |
| chr20_-_7079663 | 0.21 |
ENSDART00000040793
|
sirt5
|
sirtuin 5 |
| chr21_-_40759739 | 0.21 |
ENSDART00000075808
|
apbb3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr18_+_16726839 | 0.21 |
ENSDART00000166849
|
eif4g2b
|
eukaryotic translation initiation factor 4, gamma 2b |
| chr16_-_16329761 | 0.21 |
ENSDART00000111912
|
rbm12b
|
RNA binding motif protein 12B |
| chr20_-_7079708 | 0.20 |
ENSDART00000040793
|
sirt5
|
sirtuin 5 |
| chr8_+_54034431 | 0.20 |
ENSDART00000170712
ENSDART00000164153 |
BRPF1
|
bromodomain and PHD finger containing 1 |
| chr16_+_42567668 | 0.20 |
ENSDART00000166640
|
si:ch211-215k15.5
|
si:ch211-215k15.5 |
| chr10_-_1669268 | 0.20 |
ENSDART00000125188
|
srsf9
|
serine/arginine-rich splicing factor 9 |
| chr9_-_55250643 | 0.20 |
ENSDART00000085693
|
gpm6bb
|
glycoprotein M6Bb |
| chr19_-_30265997 | 0.20 |
ENSDART00000024292
|
txlna
|
taxilin alpha |
| chr6_-_8009055 | 0.20 |
ENSDART00000151358
|
rgl3a
|
ral guanine nucleotide dissociation stimulator-like 3a |
| chr23_-_29897606 | 0.20 |
ENSDART00000020616
|
zgc:194189
|
zgc:194189 |
| chr25_+_28380674 | 0.20 |
ENSDART00000067072
|
nfybb
|
nuclear transcription factor Y, beta b |
| chr5_+_41463969 | 0.19 |
ENSDART00000035235
|
UBB
|
si:ch211-202a12.4 |
| chr9_-_34145854 | 0.19 |
|
|
|
| chr4_+_90885 | 0.19 |
ENSDART00000169805
|
eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr1_+_30840735 | 0.19 |
ENSDART00000075286
|
slc2a15b
|
solute carrier family 2 (facilitated glucose transporter), member 15b |
| chr12_+_2769741 | 0.19 |
ENSDART00000178262
|
mms19
|
MMS19 homolog, cytosolic iron-sulfur assembly component |
| chr12_-_5153412 | 0.19 |
ENSDART00000160037
|
fra10ac1
|
FRA10A associated CGG repeat 1 |
| chr6_-_53333532 | 0.18 |
ENSDART00000172465
|
gnb1b
|
guanine nucleotide binding protein (G protein), beta polypeptide 1b |
| chr20_-_25731057 | 0.18 |
|
|
|
| chr21_-_35806609 | 0.18 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr16_-_33151504 | 0.17 |
ENSDART00000145055
|
pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr6_+_21896033 | 0.17 |
ENSDART00000163394
|
dgke
|
diacylglycerol kinase, epsilon |
| chr18_-_36291663 | 0.17 |
|
|
|
| chr8_-_20830316 | 0.17 |
ENSDART00000147267
|
si:ch211-133l5.8
|
si:ch211-133l5.8 |
| chr25_-_3377472 | 0.17 |
ENSDART00000173269
|
PRKAR2B
|
protein kinase cAMP-dependent type II regulatory subunit beta |
| chr16_+_26858600 | 0.17 |
ENSDART00000138496
|
rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr8_+_4747546 | 0.16 |
ENSDART00000045533
|
tmem127
|
transmembrane protein 127 |
| chr9_+_34623833 | 0.16 |
ENSDART00000175455
|
si:ch211-269e2.1
|
si:ch211-269e2.1 |
| chr2_-_33693243 | 0.16 |
ENSDART00000141192
|
b4galt2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr23_+_35984969 | 0.16 |
ENSDART00000130260
|
hoxc10a
|
homeobox C10a |
| chr2_+_38019291 | 0.16 |
ENSDART00000139564
|
dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr12_+_19198701 | 0.16 |
ENSDART00000100075
|
tmem184ba
|
transmembrane protein 184ba |
| chr10_+_32114762 | 0.16 |
ENSDART00000175047
|
prkrira
|
protein-kinase, interferon-inducible double stranded RNA dependent inhibitor, repressor of (P58 repressor) a |
| chr7_-_41232765 | 0.16 |
ENSDART00000173577
|
ENSDARG00000105669
|
ENSDARG00000105669 |
| chr5_-_22098977 | 0.15 |
ENSDART00000011699
|
nono
|
non-POU domain containing, octamer-binding |
| chr14_-_17270741 | 0.15 |
ENSDART00000161355
ENSDART00000168959 |
rnf4
|
ring finger protein 4 |
| chr12_+_26970745 | 0.15 |
ENSDART00000153426
|
srcap
|
Snf2-related CREBBP activator protein |
| chr3_+_34856462 | 0.15 |
ENSDART00000121981
|
smarce1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
| chr22_+_30386668 | 0.14 |
ENSDART00000059923
|
mxi1
|
max interactor 1, dimerization protein |
| chr12_-_28732946 | 0.14 |
ENSDART00000153200
|
si:ch211-194k22.8
|
si:ch211-194k22.8 |
| chr2_-_23310316 | 0.14 |
ENSDART00000146014
|
sap130b
|
Sin3A-associated protein b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0032024 | positive regulation of peptide secretion(GO:0002793) positive regulation of insulin secretion(GO:0032024) positive regulation of hormone secretion(GO:0046887) positive regulation of peptide hormone secretion(GO:0090277) |
| 0.2 | 1.2 | GO:0055129 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 2.0 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.2 | 0.7 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.2 | 0.6 | GO:0010482 | keratinocyte development(GO:0003334) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.2 | 4.4 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.1 | 2.8 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.1 | 0.7 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 1.3 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.4 | GO:0036047 | regulation of ketone biosynthetic process(GO:0010566) protein demalonylation(GO:0036046) peptidyl-lysine demalonylation(GO:0036047) protein desuccinylation(GO:0036048) peptidyl-lysine desuccinylation(GO:0036049) protein deglutarylation(GO:0061698) |
| 0.1 | 0.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.3 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.1 | 0.3 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 0.2 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.1 | 0.2 | GO:0051580 | neurotransmitter uptake(GO:0001504) regulation of neurotransmitter uptake(GO:0051580) neurotransmitter reuptake(GO:0098810) |
| 0.1 | 0.3 | GO:2000001 | regulation of DNA damage checkpoint(GO:2000001) |
| 0.1 | 0.2 | GO:0021547 | midbrain-hindbrain boundary initiation(GO:0021547) |
| 0.1 | 0.4 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 0.2 | GO:0051660 | neuroblast proliferation(GO:0007405) establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.2 | GO:1905168 | positive regulation of double-strand break repair via homologous recombination(GO:1905168) |
| 0.0 | 0.9 | GO:0006625 | protein targeting to peroxisome(GO:0006625) peroxisomal transport(GO:0043574) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 2.0 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 0.8 | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) negative regulation of proteasomal protein catabolic process(GO:1901799) negative regulation of proteolysis involved in cellular protein catabolic process(GO:1903051) |
| 0.0 | 0.2 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.7 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.0 | 0.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.3 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:1902946 | protein localization to early endosome(GO:1902946) |
| 0.0 | 0.9 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.0 | GO:0008643 | carbohydrate transport(GO:0008643) |
| 0.0 | 0.2 | GO:0007205 | protein kinase C-activating G-protein coupled receptor signaling pathway(GO:0007205) |
| 0.0 | 0.3 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 1.1 | GO:1902593 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.1 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.0 | 0.6 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.0 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.9 | GO:0071599 | otic vesicle development(GO:0071599) |
| 0.0 | 0.1 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.9 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.5 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.5 | GO:0070304 | positive regulation of stress-activated MAPK cascade(GO:0032874) positive regulation of stress-activated protein kinase signaling cascade(GO:0070304) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.1 | 2.8 | GO:0008328 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.1 | 1.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.2 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 2.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.1 | 0.8 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.0 | 0.7 | GO:0000177 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 1.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:0043189 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 5.5 | GO:0005912 | adherens junction(GO:0005912) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.5 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.7 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.9 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.2 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 1.5 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.5 | GO:0016459 | myosin complex(GO:0016459) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.2 | 0.8 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.2 | 0.6 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 2.0 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.6 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.4 | GO:0036054 | protein-malonyllysine demalonylase activity(GO:0036054) protein-succinyllysine desuccinylase activity(GO:0036055) protein-glutaryllysine deglutarylase activity(GO:0061697) |
| 0.1 | 0.4 | GO:0005165 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.1 | 4.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.4 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.6 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 1.0 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 0.4 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 1.2 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.3 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.8 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.0 | 0.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.0 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 2.4 | GO:0003713 | transcription coactivator activity(GO:0003713) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.4 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 2.8 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.1 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.0 | 1.3 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.0 | GO:0032135 | DNA insertion or deletion binding(GO:0032135) |
| 0.0 | 0.1 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 0.9 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 2.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.3 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.4 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.2 | 4.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 1.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.1 | 0.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.7 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.7 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |