Project
DANIO-CODE
Navigation
Downloads
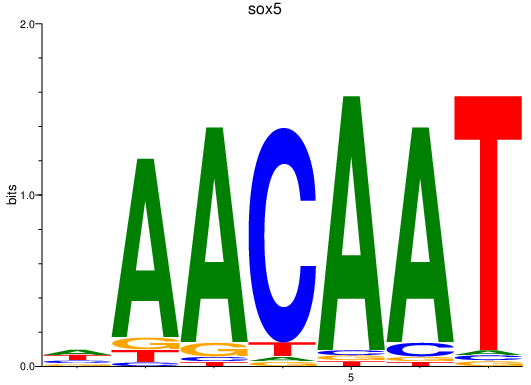
Results for sox5
Z-value: 2.57
Transcription factors associated with sox5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox5
|
ENSDARG00000011582 | SRY-box transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox5 | dr10_dc_chr4_-_17066886_17066993 | 0.98 | 9.8e-12 | Click! |
Activity profile of sox5 motif
Sorted Z-values of sox5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of sox5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_28753020 | 7.83 |
ENSDART00000155815
|
nova2
|
neuro-oncological ventral antigen 2 |
| chr22_-_37417903 | 7.06 |
ENSDART00000149948
|
FP102167.1
|
ENSDARG00000095844 |
| chr8_-_40231417 | 6.63 |
ENSDART00000162020
|
kdm2ba
|
lysine (K)-specific demethylase 2Ba |
| chr5_+_34857986 | 6.61 |
ENSDART00000141239
|
erlin2
|
ER lipid raft associated 2 |
| chr10_+_33227967 | 6.27 |
ENSDART00000159666
|
myl10
|
myosin, light chain 10, regulatory |
| chr8_+_21163236 | 5.82 |
ENSDART00000091307
|
col2a1a
|
collagen, type II, alpha 1a |
| chr8_-_16223730 | 5.52 |
ENSDART00000057590
|
dmrta2
|
DMRT-like family A2 |
| chr24_-_24306469 | 5.07 |
ENSDART00000154149
|
BX323067.1
|
ENSDARG00000097984 |
| chr7_+_47014195 | 4.98 |
ENSDART00000114669
|
dpy19l3
|
dpy-19-like 3 (C. elegans) |
| chr16_-_7525980 | 4.94 |
ENSDART00000017445
|
prdm1a
|
PR domain containing 1a, with ZNF domain |
| chr14_+_38445969 | 4.86 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr18_+_16257606 | 4.78 |
ENSDART00000142584
|
alx1
|
ALX homeobox 1 |
| chr22_-_27082972 | 4.68 |
ENSDART00000077411
|
cxcl12b
|
chemokine (C-X-C motif) ligand 12b (stromal cell-derived factor 1) |
| chr20_-_35343242 | 4.62 |
ENSDART00000113294
|
fzd3a
|
frizzled class receptor 3a |
| chr1_-_34921848 | 4.61 |
ENSDART00000142154
|
frem3
|
Fras1 related extracellular matrix 3 |
| chr5_+_6054781 | 4.61 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr7_-_50827308 | 4.56 |
ENSDART00000121574
|
col4a6
|
collagen, type IV, alpha 6 |
| chr17_-_39237265 | 4.54 |
ENSDART00000050534
|
crim1
|
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
| chr11_-_41357639 | 4.50 |
ENSDART00000055709
|
her2
|
hairy-related 2 |
| chr19_-_22184035 | 4.50 |
|
|
|
| chr24_-_6617860 | 4.36 |
ENSDART00000166216
|
arhgap21a
|
Rho GTPase activating protein 21a |
| chr13_-_22641001 | 4.36 |
|
|
|
| chr21_-_35806638 | 4.24 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr23_-_15442032 | 4.21 |
ENSDART00000082060
|
ENSDARG00000078145
|
ENSDARG00000078145 |
| chr18_+_40364675 | 4.18 |
ENSDART00000098791
ENSDART00000049171 |
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr24_+_9605832 | 4.16 |
ENSDART00000131891
|
tmem108
|
transmembrane protein 108 |
| chr25_+_14411153 | 4.03 |
ENSDART00000015681
|
dbx1b
|
developing brain homeobox 1b |
| chr5_+_17120453 | 4.01 |
|
|
|
| chr7_-_38590391 | 3.99 |
ENSDART00000037361
ENSDART00000173629 ENSDART00000173953 |
phf21aa
|
PHD finger protein 21Aa |
| chr6_+_47845002 | 3.99 |
ENSDART00000140943
|
padi2
|
peptidyl arginine deiminase, type II |
| chr9_-_34129128 | 3.91 |
|
|
|
| chr5_-_24509021 | 3.87 |
ENSDART00000177338
ENSDART00000177557 ENSDART00000177478 |
emid1
|
EMI domain containing 1 |
| chr13_+_11913290 | 3.82 |
ENSDART00000079398
|
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr14_-_26406720 | 3.79 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr14_+_34683602 | 3.77 |
ENSDART00000172171
|
ebf1a
|
early B-cell factor 1a |
| chr21_+_28465617 | 3.73 |
ENSDART00000140229
|
otub1a
|
OTU deubiquitinase, ubiquitin aldehyde binding 1a |
| chr16_+_28819826 | 3.69 |
ENSDART00000103340
|
s100v1
|
S100 calcium binding protein V1 |
| chr6_+_42821679 | 3.67 |
ENSDART00000046498
|
sema3fa
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3Fa |
| chr11_+_27027209 | 3.63 |
ENSDART00000113707
|
fbln2
|
fibulin 2 |
| chr13_-_36996246 | 3.62 |
ENSDART00000133242
|
syne2b
|
spectrin repeat containing, nuclear envelope 2b |
| chr13_-_22901327 | 3.61 |
ENSDART00000056523
|
hkdc1
|
hexokinase domain containing 1 |
| chr3_-_31672763 | 3.49 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr16_+_23172295 | 3.45 |
ENSDART00000167518
ENSDART00000161087 |
efna3b
|
ephrin-A3b |
| chr8_+_23071884 | 3.44 |
ENSDART00000063075
|
zgc:100920
|
zgc:100920 |
| chr21_-_25704662 | 3.27 |
ENSDART00000101211
|
cldnh
|
claudin h |
| chr13_-_24779651 | 3.27 |
ENSDART00000077775
|
kat6b
|
K(lysine) acetyltransferase 6B |
| chr21_+_20734431 | 3.25 |
ENSDART00000079732
|
oxct1b
|
3-oxoacid CoA transferase 1b |
| chr9_-_22528568 | 3.21 |
ENSDART00000134805
|
crygm2d1
|
crystallin, gamma M2d1 |
| chr13_+_11912981 | 3.21 |
ENSDART00000158244
|
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr11_-_23206021 | 3.20 |
ENSDART00000032844
|
plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr19_+_48582114 | 3.19 |
ENSDART00000157424
|
CU693379.1
|
ENSDARG00000098777 |
| chr10_+_21850059 | 3.18 |
ENSDART00000164634
ENSDART00000172513 |
pcdh1g32
|
protocadherin 1 gamma 32 |
| chr13_-_25067585 | 3.11 |
ENSDART00000159585
|
adka
|
adenosine kinase a |
| chr23_-_18972097 | 3.10 |
ENSDART00000133826
|
CR847953.1
|
ENSDARG00000057403 |
| chr16_+_5625301 | 3.09 |
|
|
|
| chr13_+_11305846 | 3.08 |
|
|
|
| chr19_-_7069920 | 3.07 |
ENSDART00000145741
|
znf384l
|
zinc finger protein 384 like |
| chr3_-_30027552 | 3.04 |
ENSDART00000103502
|
si:ch211-152f23.5
|
si:ch211-152f23.5 |
| chr1_-_37990863 | 3.02 |
ENSDART00000132402
|
gpm6ab
|
glycoprotein M6Ab |
| chr25_+_24152717 | 3.01 |
ENSDART00000064646
|
tmem86a
|
transmembrane protein 86A |
| chr16_+_29558320 | 2.98 |
ENSDART00000011497
|
ctsk
|
cathepsin K |
| chr9_-_31467299 | 2.96 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr7_-_58427369 | 2.91 |
ENSDART00000149347
|
chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr19_-_34508557 | 2.91 |
ENSDART00000160495
|
si:dkey-184p18.2
|
si:dkey-184p18.2 |
| chr12_-_26314881 | 2.86 |
ENSDART00000178687
|
myoz1b
|
myozenin 1b |
| chr15_-_20297270 | 2.82 |
ENSDART00000123910
|
ppp1r14ab
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Ab |
| chr23_-_10747605 | 2.81 |
|
|
|
| chr2_+_22196971 | 2.78 |
ENSDART00000140012
|
ca8
|
carbonic anhydrase VIII |
| chr6_-_44282665 | 2.78 |
ENSDART00000157215
|
pdzrn3b
|
PDZ domain containing RING finger 3b |
| chr24_+_21395671 | 2.76 |
ENSDART00000091529
|
wasf3b
|
WAS protein family, member 3b |
| chr20_-_47521258 | 2.74 |
|
|
|
| chr23_+_35984675 | 2.74 |
ENSDART00000053295
|
hoxc10a
|
homeobox C10a |
| chr11_-_25222937 | 2.69 |
|
|
|
| chr19_-_2486568 | 2.68 |
ENSDART00000012791
|
sp8a
|
sp8 transcription factor a |
| chr18_-_14868444 | 2.66 |
ENSDART00000045232
|
mtss1la
|
metastasis suppressor 1-like a |
| chr25_-_20994084 | 2.64 |
ENSDART00000154765
|
prr5a
|
proline rich 5a (renal) |
| chr7_-_52283383 | 2.62 |
ENSDART00000165649
|
tcf12
|
transcription factor 12 |
| chr7_-_33080261 | 2.58 |
ENSDART00000114041
|
anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr24_-_20513975 | 2.58 |
ENSDART00000127923
|
nktr
|
natural killer cell triggering receptor |
| chr16_+_10427961 | 2.57 |
ENSDART00000055380
|
tubb5
|
tubulin, beta 5 |
| chr11_-_42934175 | 2.57 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr7_+_65532390 | 2.57 |
ENSDART00000156683
|
CT573494.2
|
ENSDARG00000097673 |
| chr6_-_8075384 | 2.56 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr16_+_19926776 | 2.56 |
ENSDART00000149901
ENSDART00000052927 |
twist1b
|
twist family bHLH transcription factor 1b |
| chr13_-_20387917 | 2.54 |
ENSDART00000167916
ENSDART00000165310 ENSDART00000168955 |
gfra1a
|
gdnf family receptor alpha 1a |
| chr10_+_31358236 | 2.54 |
ENSDART00000145562
|
robo4
|
roundabout, axon guidance receptor, homolog 4 (Drosophila) |
| chr9_+_4188263 | 2.49 |
ENSDART00000164543
|
kalrna
|
kalirin RhoGEF kinase a |
| chr8_-_40231354 | 2.48 |
ENSDART00000162020
|
kdm2ba
|
lysine (K)-specific demethylase 2Ba |
| chr1_+_8605984 | 2.46 |
ENSDART00000055009
|
uncx4.1
|
Unc4.1 homeobox (C. elegans) |
| chr24_-_36938495 | 2.45 |
ENSDART00000112694
|
fam171a2b
|
family with sequence similarity 171, member A2b |
| chr23_-_5751064 | 2.44 |
ENSDART00000067351
|
tnnt2a
|
troponin T type 2a (cardiac) |
| chr18_+_40365208 | 2.42 |
ENSDART00000167134
|
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr24_-_6048914 | 2.38 |
ENSDART00000146830
ENSDART00000021981 |
apbb1ip
|
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
| chr9_+_33167554 | 2.38 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr12_-_1931281 | 2.36 |
ENSDART00000005676
ENSDART00000127937 |
sox9a
|
SRY (sex determining region Y)-box 9a |
| chr11_-_6051096 | 2.35 |
ENSDART00000147761
|
vsg1
|
vessel-specific 1 |
| chr19_+_44373777 | 2.35 |
ENSDART00000051712
|
gatad1
|
GATA zinc finger domain containing 1 |
| chr19_+_5399813 | 2.33 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr20_+_20287015 | 2.32 |
ENSDART00000002507
|
rhoj
|
ras homolog family member J |
| chr7_+_26980284 | 2.29 |
ENSDART00000173962
|
sox6
|
SRY (sex determining region Y)-box 6 |
| chr1_-_29885008 | 2.29 |
ENSDART00000018827
|
dachc
|
dachshund c |
| chr2_-_50638153 | 2.27 |
ENSDART00000108900
|
cntnap2b
|
contactin associated protein-like 2b |
| chr23_+_20936419 | 2.27 |
ENSDART00000129992
|
pax7b
|
paired box 7b |
| chr5_-_40310798 | 2.27 |
ENSDART00000133183
|
parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr19_-_32931355 | 2.24 |
ENSDART00000103410
|
zbtb8b
|
zinc finger and BTB domain containing 8B |
| chr1_-_15972040 | 2.24 |
ENSDART00000040434
|
asah1b
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1b |
| chr13_+_1051015 | 2.24 |
ENSDART00000033528
|
tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr17_-_52735250 | 2.22 |
|
|
|
| chr6_+_43429115 | 2.20 |
ENSDART00000056457
|
mitfa
|
microphthalmia-associated transcription factor a |
| chr22_-_15576473 | 2.19 |
ENSDART00000036075
|
tpm4a
|
tropomyosin 4a |
| chr17_-_1599376 | 2.16 |
ENSDART00000034216
ENSDART00000158837 |
dync1h1
|
dynein, cytoplasmic 1, heavy chain 1 |
| chr10_+_33935945 | 2.15 |
|
|
|
| chr17_+_34854673 | 2.15 |
|
|
|
| chr4_-_6558919 | 2.12 |
ENSDART00000142087
|
foxp2
|
forkhead box P2 |
| chr10_+_38582701 | 2.07 |
ENSDART00000144329
|
acer3
|
alkaline ceramidase 3 |
| chr14_+_34683695 | 2.04 |
ENSDART00000170631
|
ebf1a
|
early B-cell factor 1a |
| chr8_+_23704956 | 2.03 |
ENSDART00000132734
|
mkrn4
|
makorin, ring finger protein, 4 |
| chr11_+_6809190 | 2.03 |
ENSDART00000104289
|
rab3ab
|
RAB3A, member RAS oncogene family, b |
| chr19_-_31996936 | 2.02 |
ENSDART00000052183
|
zgc:111986
|
zgc:111986 |
| chr19_-_31754505 | 2.02 |
ENSDART00000040810
|
arl4ab
|
ADP-ribosylation factor-like 4ab |
| chr11_+_37345024 | 1.97 |
ENSDART00000077496
|
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr5_-_40310638 | 1.96 |
ENSDART00000133183
|
parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr7_-_52574822 | 1.95 |
ENSDART00000172951
|
map1aa
|
microtubule-associated protein 1Aa |
| chr9_+_24198112 | 1.92 |
ENSDART00000148226
|
mlphb
|
melanophilin b |
| chr9_-_10561062 | 1.91 |
ENSDART00000175269
|
thsd7ba
|
thrombospondin, type I, domain containing 7Ba |
| chr4_-_17960341 | 1.90 |
ENSDART00000144956
|
anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| KN149760v1_-_29067 | 1.89 |
|
|
|
| chr23_+_18796386 | 1.88 |
ENSDART00000137438
|
myh7bb
|
myosin, heavy chain 7B, cardiac muscle, beta b |
| chr23_+_20936374 | 1.88 |
ENSDART00000129992
|
pax7b
|
paired box 7b |
| chr13_+_30190977 | 1.87 |
|
|
|
| chr3_+_47826386 | 1.87 |
ENSDART00000003338
|
unkl
|
unkempt family zinc finger-like |
| chr8_+_20456215 | 1.85 |
ENSDART00000036630
|
zgc:101100
|
zgc:101100 |
| chr20_-_35343057 | 1.84 |
ENSDART00000113294
|
fzd3a
|
frizzled class receptor 3a |
| chr22_-_20317201 | 1.84 |
ENSDART00000161610
|
tcf3b
|
transcription factor 3b |
| chr21_+_17073163 | 1.83 |
|
|
|
| chr13_+_30546398 | 1.83 |
|
|
|
| chr6_-_41087828 | 1.81 |
ENSDART00000028217
|
srsf3a
|
serine/arginine-rich splicing factor 3a |
| chr3_+_52745202 | 1.81 |
ENSDART00000104683
|
pbx4
|
pre-B-cell leukemia transcription factor 4 |
| chr25_-_28691412 | 1.79 |
ENSDART00000067066
|
parp6b
|
poly (ADP-ribose) polymerase family, member 6b |
| chr21_-_27164751 | 1.79 |
ENSDART00000065402
|
zgc:77262
|
zgc:77262 |
| chr8_-_12171624 | 1.78 |
ENSDART00000132824
|
dab2ipa
|
DAB2 interacting protein a |
| chr8_+_7655655 | 1.78 |
ENSDART00000170184
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr19_-_7069850 | 1.77 |
ENSDART00000145741
|
znf384l
|
zinc finger protein 384 like |
| chr21_-_20728623 | 1.74 |
ENSDART00000135940
ENSDART00000002231 |
ghrb
|
growth hormone receptor b |
| chr1_+_45102814 | 1.71 |
|
|
|
| chr2_+_22196842 | 1.69 |
ENSDART00000140012
|
ca8
|
carbonic anhydrase VIII |
| chr8_+_39525254 | 1.67 |
|
|
|
| chr16_-_28921444 | 1.66 |
ENSDART00000078543
|
syt11b
|
synaptotagmin XIb |
| chr2_+_3125365 | 1.66 |
ENSDART00000164308
ENSDART00000167649 |
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| KN150129v1_-_30546 | 1.66 |
|
|
|
| chr24_-_28166614 | 1.65 |
ENSDART00000167861
|
cox4i1l
|
cytochrome c oxidase subunit IV isoform 1, like |
| chr17_-_52735615 | 1.65 |
|
|
|
| chr17_+_28690237 | 1.64 |
ENSDART00000126967
|
strn3
|
striatin, calmodulin binding protein 3 |
| chr5_-_40310702 | 1.64 |
ENSDART00000133183
|
parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr7_-_68033217 | 1.62 |
|
|
|
| chr23_-_3816492 | 1.59 |
ENSDART00000136394
|
hmga1a
|
high mobility group AT-hook 1a |
| chr17_-_21422311 | 1.59 |
ENSDART00000167094
|
vax1
|
ventral anterior homeobox 1 |
| chr11_-_3846064 | 1.55 |
ENSDART00000161426
|
gata2a
|
GATA binding protein 2a |
| chr25_-_36760720 | 1.51 |
ENSDART00000111862
|
ldlrad3
|
low density lipoprotein receptor class A domain containing 3 |
| chr19_+_20189224 | 1.51 |
ENSDART00000163611
|
hoxa4a
|
homeobox A4a |
| chr9_-_22371512 | 1.51 |
ENSDART00000101809
|
crygm2d6
|
crystallin, gamma M2d6 |
| chr9_+_49961721 | 1.49 |
|
|
|
| chr24_-_8589800 | 1.47 |
|
|
|
| chr22_-_20316908 | 1.47 |
ENSDART00000165667
|
tcf3b
|
transcription factor 3b |
| chr13_-_8268681 | 1.46 |
ENSDART00000045086
|
prkceb
|
protein kinase C, epsilon b |
| chr11_-_30971376 | 1.45 |
ENSDART00000170700
|
nacc1b
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing b |
| chr17_+_19003200 | 1.42 |
|
|
|
| chr11_-_40415291 | 1.41 |
|
|
|
| chr19_+_5399877 | 1.41 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr25_+_7147666 | 1.40 |
ENSDART00000104712
|
hmg20a
|
high mobility group 20A |
| chr11_+_26138359 | 1.39 |
ENSDART00000087652
|
cpne1
|
copine I |
| chr6_-_43679553 | 1.39 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr18_+_14725148 | 1.38 |
ENSDART00000146128
|
uri1
|
URI1, prefoldin-like chaperone |
| chr21_+_27241416 | 1.37 |
|
|
|
| chr14_-_30467807 | 1.36 |
ENSDART00000173262
|
prss23
|
protease, serine, 23 |
| chr9_-_22324699 | 1.36 |
ENSDART00000175417
ENSDART00000101902 |
crygm2d8
|
crystallin, gamma M2d8 |
| chr5_-_45219030 | 1.36 |
|
|
|
| chr17_-_35330764 | 1.35 |
ENSDART00000063437
|
adam17a
|
ADAM metallopeptidase domain 17a |
| chr23_-_33728344 | 1.34 |
ENSDART00000146180
|
csrnp2
|
cysteine-serine-rich nuclear protein 2 |
| chr5_+_64159481 | 1.33 |
ENSDART00000073953
|
lrrc8ab
|
leucine rich repeat containing 8 family, member Ab |
| chr21_-_25704793 | 1.31 |
ENSDART00000101211
|
cldnh
|
claudin h |
| chr8_+_13069277 | 1.31 |
ENSDART00000138433
|
itgb4
|
integrin, beta 4 |
| chr5_+_32680990 | 1.30 |
ENSDART00000146223
|
surf1
|
surfeit 1 |
| chr18_+_6692267 | 1.30 |
|
|
|
| chr23_+_35819758 | 1.30 |
ENSDART00000049551
|
rarga
|
retinoic acid receptor gamma a |
| chr3_-_41944723 | 1.30 |
ENSDART00000083111
|
ttyh3a
|
tweety family member 3a |
| chr2_+_31974269 | 1.27 |
ENSDART00000012413
|
ankhb
|
ANKH inorganic pyrophosphate transport regulator b |
| chr23_-_32231333 | 1.27 |
ENSDART00000000876
|
nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr3_+_24003840 | 1.26 |
ENSDART00000156204
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr13_-_20388016 | 1.25 |
ENSDART00000167916
ENSDART00000165310 ENSDART00000168955 |
gfra1a
|
gdnf family receptor alpha 1a |
| chr3_-_35472632 | 1.25 |
ENSDART00000055269
|
gng13b
|
guanine nucleotide binding protein (G protein), gamma 13b |
| chr13_-_40286013 | 1.24 |
ENSDART00000150091
ENSDART00000057094 |
nkx2.3
|
NK2 homeobox 3 |
| chr4_-_6558687 | 1.24 |
ENSDART00000150774
|
foxp2
|
forkhead box P2 |
| chr5_+_61335845 | 1.24 |
ENSDART00000166482
ENSDART00000050884 ENSDART00000163909 |
sepw1
|
selenoprotein W, 1 |
| chr17_-_33336706 | 1.19 |
ENSDART00000040346
|
efr3ba
|
EFR3 homolog Ba (S. cerevisiae) |
| chr12_+_26530620 | 1.19 |
ENSDART00000046959
|
arhgap12b
|
Rho GTPase activating protein 12b |
| chr8_+_30654957 | 1.18 |
ENSDART00000138750
|
adora2aa
|
adenosine A2a receptor a |
| chr13_+_22165699 | 1.17 |
|
|
|
| chr25_-_20993989 | 1.15 |
|
|
|
| chr23_+_36019989 | 1.15 |
ENSDART00000103134
ENSDART00000139319 |
hoxc5a
|
homeobox C5a |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.8 | 7.1 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 1.6 | 4.8 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 1.4 | 4.2 | GO:0097484 | dendrite extension(GO:0097484) |
| 1.4 | 5.5 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 1.3 | 5.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 1.2 | 4.7 | GO:0050938 | regulation of xanthophore differentiation(GO:0050938) |
| 0.9 | 3.8 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.9 | 4.7 | GO:0032332 | positive regulation of chondrocyte differentiation(GO:0032332) |
| 0.8 | 3.2 | GO:0046950 | cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.8 | 3.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.8 | 4.5 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.7 | 2.2 | GO:0071947 | protein K29-linked deubiquitination(GO:0035523) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.6 | 6.5 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.6 | 6.6 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.5 | 4.9 | GO:0051155 | positive regulation of striated muscle cell differentiation(GO:0051155) |
| 0.5 | 3.7 | GO:1901315 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.5 | 1.5 | GO:1902893 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.5 | 2.0 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.5 | 1.4 | GO:0001961 | positive regulation of cytokine-mediated signaling pathway(GO:0001961) positive regulation of response to cytokine stimulus(GO:0060760) |
| 0.4 | 2.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.4 | 2.6 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.4 | 7.7 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.4 | 3.4 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.4 | 1.1 | GO:0086010 | membrane depolarization during action potential(GO:0086010) |
| 0.4 | 3.0 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.4 | 1.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 4.4 | GO:0045471 | response to ethanol(GO:0045471) |
| 0.3 | 8.1 | GO:0008214 | protein demethylation(GO:0006482) protein dealkylation(GO:0008214) |
| 0.3 | 1.0 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.3 | 5.9 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.3 | 1.8 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.3 | 1.8 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.3 | 5.5 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.3 | 2.6 | GO:0090259 | regulation of retinal ganglion cell axon guidance(GO:0090259) |
| 0.3 | 4.5 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.3 | 7.0 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.3 | 0.8 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.3 | 4.6 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.2 | 1.7 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.2 | 1.0 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.2 | 2.8 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.2 | 2.1 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.2 | 1.4 | GO:0003160 | endocardium morphogenesis(GO:0003160) |
| 0.2 | 3.8 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.2 | 1.2 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.2 | 2.7 | GO:0097178 | ruffle assembly(GO:0097178) |
| 0.2 | 1.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.2 | 5.0 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.2 | 3.7 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.2 | 1.0 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.2 | 1.7 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.2 | 4.2 | GO:0015696 | ammonium transport(GO:0015696) |
| 0.2 | 1.3 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.2 | 2.2 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.2 | 1.8 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.1 | 3.4 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.1 | 6.8 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of axon guidance(GO:1902668) |
| 0.1 | 0.8 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 0.1 | 1.0 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 2.0 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
| 0.1 | 8.6 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 5.9 | GO:0021515 | cell differentiation in spinal cord(GO:0021515) |
| 0.1 | 1.7 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 3.0 | GO:0009612 | response to mechanical stimulus(GO:0009612) |
| 0.1 | 3.8 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.1 | 2.6 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.9 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.1 | 2.7 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.1 | 0.8 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 6.1 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 1.8 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 2.6 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.1 | 2.8 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.1 | 0.3 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.1 | 0.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 4.9 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 0.8 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.1 | 1.3 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.1 | 2.2 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.1 | 0.5 | GO:0042551 | neuron maturation(GO:0042551) |
| 0.1 | 0.6 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 1.1 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 1.5 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.1 | 2.5 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.1 | 0.5 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 1.8 | GO:0043242 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) negative regulation of protein complex disassembly(GO:0043242) actin filament capping(GO:0051693) negative regulation of protein depolymerization(GO:1901880) |
| 0.0 | 4.5 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 1.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.3 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.8 | GO:0030032 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.0 | 0.4 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 1.2 | GO:0048546 | digestive tract morphogenesis(GO:0048546) |
| 0.0 | 0.2 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 2.6 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.3 | GO:0051932 | synaptic transmission, GABAergic(GO:0051932) regulation of postsynaptic membrane potential(GO:0060078) |
| 0.0 | 0.1 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 3.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.3 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.5 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 0.2 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) |
| 0.0 | 0.5 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.8 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 1.9 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 1.3 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 1.1 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 0.5 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.3 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.6 | GO:0007254 | JNK cascade(GO:0007254) |
| 0.0 | 2.0 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.5 | GO:1901653 | cellular response to peptide hormone stimulus(GO:0071375) cellular response to peptide(GO:1901653) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 4.6 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.6 | 4.5 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.6 | 7.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.6 | 1.7 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.6 | 3.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.4 | 1.8 | GO:0008091 | spectrin(GO:0008091) |
| 0.3 | 0.8 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 1.6 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.2 | 2.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 7.3 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 3.6 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.2 | 5.5 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.2 | 1.1 | GO:0034706 | voltage-gated sodium channel complex(GO:0001518) sodium channel complex(GO:0034706) |
| 0.1 | 5.1 | GO:0044420 | basement membrane(GO:0005604) extracellular matrix component(GO:0044420) |
| 0.1 | 0.5 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 2.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 5.9 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 1.0 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 2.2 | GO:0030286 | dynein complex(GO:0030286) |
| 0.1 | 4.2 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.1 | 5.4 | GO:0031674 | I band(GO:0031674) |
| 0.1 | 0.6 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.1 | 1.6 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 1.8 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 2.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 4.0 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 3.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.8 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 2.0 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.0 | 4.5 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.0 | 5.0 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 4.1 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 1.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.3 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 8.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 2.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 5.9 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 1.3 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 13.3 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 2.0 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 6.9 | GO:0005789 | endoplasmic reticulum membrane(GO:0005789) |
| 0.0 | 0.5 | GO:0042383 | sarcolemma(GO:0042383) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 5.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 1.0 | 3.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.9 | 4.3 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.8 | 3.2 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.8 | 3.0 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.7 | 2.9 | GO:0031433 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.7 | 8.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.6 | 1.7 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.5 | 3.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.5 | 4.7 | GO:0008009 | chemokine activity(GO:0008009) chemokine receptor binding(GO:0042379) |
| 0.5 | 2.6 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.5 | 3.7 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.4 | 2.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.4 | 1.3 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.4 | 1.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.3 | 6.5 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.3 | 4.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.3 | 1.8 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.3 | 2.8 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.3 | 2.4 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.3 | 3.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.3 | 0.8 | GO:0008126 | acetylesterase activity(GO:0008126) |
| 0.3 | 7.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.3 | 1.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 1.3 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.2 | 3.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 4.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 8.1 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.2 | 2.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 6.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 4.5 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.2 | 11.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.2 | 3.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.2 | 3.0 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 1.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 5.5 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.2 | 6.6 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.2 | 2.2 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 2.6 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.2 | 1.1 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.1 | 0.6 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 4.9 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.1 | 4.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 6.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 4.4 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.4 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.1 | 1.7 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 1.4 | GO:0004697 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.1 | 0.3 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.1 | 3.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.8 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 2.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.5 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 1.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 1.0 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 1.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 1.1 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) phospholipase C activity(GO:0004629) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.5 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 1.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 44.0 | GO:0001159 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 1.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 4.8 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 11.9 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.8 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.7 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 0.2 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 1.1 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.9 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 1.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 4.5 | GO:0005543 | phospholipid binding(GO:0005543) |
| 0.0 | 8.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 5.6 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 5.6 | GO:1990837 | sequence-specific double-stranded DNA binding(GO:1990837) |
| 0.0 | 0.9 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 1.0 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 3.4 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 3.9 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 0.8 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.1 | GO:0003714 | transcription corepressor activity(GO:0003714) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 7.0 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.3 | 2.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 5.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.2 | 12.0 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.2 | 1.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 5.0 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 2.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 4.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 3.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 7.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.6 | 3.0 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.3 | 2.6 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.3 | 2.4 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.3 | 1.8 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 5.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 2.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 2.1 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 2.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.3 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.3 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.1 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 2.2 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 0.9 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.8 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 2.6 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |