Project
DANIO-CODE
Navigation
Downloads
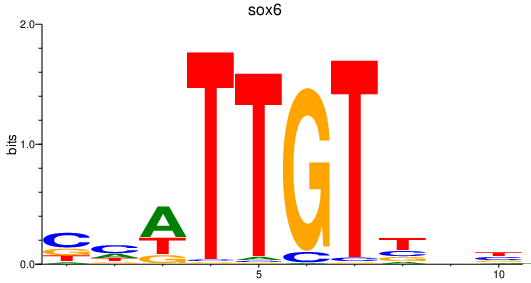
Results for sox6
Z-value: 0.59
Transcription factors associated with sox6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox6
|
ENSDARG00000015536 | SRY-box transcription factor 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox6 | dr10_dc_chr7_+_26978970_26979151 | -0.80 | 2.3e-04 | Click! |
Activity profile of sox6 motif
Sorted Z-values of sox6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of sox6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_48356793 | 2.94 |
ENSDART00000054788
ENSDART00000152899 |
ddit4
|
DNA-damage-inducible transcript 4 |
| chr21_-_2292663 | 1.69 |
ENSDART00000164015
|
zgc:66483
|
zgc:66483 |
| chr7_-_24249672 | 1.29 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr2_-_57020663 | 1.21 |
|
|
|
| chr24_-_19573966 | 1.17 |
ENSDART00000158952
ENSDART00000109107 ENSDART00000056082 |
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr2_+_32863386 | 1.14 |
ENSDART00000056649
|
tmem53
|
transmembrane protein 53 |
| chr23_-_34017934 | 1.09 |
ENSDART00000133223
|
ENSDARG00000070393
|
ENSDARG00000070393 |
| chr24_-_19574666 | 1.04 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr13_+_46652067 | 1.04 |
ENSDART00000056962
|
fbxo5
|
F-box protein 5 |
| chr2_+_27730940 | 1.02 |
ENSDART00000134976
|
si:ch73-382f3.1
|
si:ch73-382f3.1 |
| chr9_+_21982679 | 1.02 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr9_+_21982644 | 0.96 |
ENSDART00000134915
|
rev1
|
REV1, polymerase (DNA directed) |
| chr8_+_7655257 | 0.91 |
ENSDART00000170184
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr17_+_7377230 | 0.86 |
ENSDART00000157123
|
shprh
|
SNF2 histone linker PHD RING helicase |
| chr16_-_55211053 | 0.81 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr3_+_44928592 | 0.79 |
ENSDART00000168864
|
zgc:112146
|
zgc:112146 |
| chr6_+_35811398 | 0.78 |
ENSDART00000151760
|
FP236809.1
|
ENSDARG00000096356 |
| chr2_+_30800532 | 0.76 |
|
|
|
| chr3_+_44928323 | 0.75 |
ENSDART00000170913
|
zgc:112146
|
zgc:112146 |
| chr6_-_3763940 | 0.75 |
ENSDART00000171804
|
tlk1b
|
tousled-like kinase 1b |
| chr23_-_34017830 | 0.73 |
ENSDART00000133223
|
ENSDARG00000070393
|
ENSDARG00000070393 |
| chr7_-_24249630 | 0.72 |
ENSDART00000077039
|
faah2b
|
fatty acid amide hydrolase 2b |
| chr8_+_7655310 | 0.72 |
ENSDART00000170184
|
fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr2_-_41598080 | 0.70 |
ENSDART00000178357
ENSDART00000176801 ENSDART00000177840 |
CU302321.2
|
ENSDARG00000107739 |
| chr3_-_25246374 | 0.68 |
ENSDART00000077538
|
kpna2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
| chr7_-_52847458 | 0.68 |
ENSDART00000172179
ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr3_-_25246452 | 0.68 |
ENSDART00000177576
|
kpna2
|
karyopherin alpha 2 (RAG cohort 1, importin alpha 1) |
| chr2_-_57020490 | 0.66 |
|
|
|
| chr18_-_24490329 | 0.66 |
ENSDART00000147961
|
CR847971.2
|
ENSDARG00000094853 |
| chr6_-_7577977 | 0.66 |
ENSDART00000151545
|
slc25a38b
|
solute carrier family 25, member 38b |
| chr5_+_26888419 | 0.66 |
ENSDART00000123635
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr5_-_27549500 | 0.63 |
ENSDART00000051515
|
zgc:110329
|
zgc:110329 |
| chr20_-_3301898 | 0.62 |
ENSDART00000123096
|
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr16_+_32060609 | 0.59 |
ENSDART00000047570
|
mboat7
|
membrane bound O-acyltransferase domain containing 7 |
| chr9_+_21982756 | 0.59 |
ENSDART00000059652
|
rev1
|
REV1, polymerase (DNA directed) |
| chr19_+_47835134 | 0.59 |
ENSDART00000171281
|
psmb2
|
proteasome subunit beta 2 |
| chr13_+_33238135 | 0.57 |
ENSDART00000158709
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr20_+_49332270 | 0.57 |
|
|
|
| chr13_+_33237979 | 0.56 |
ENSDART00000158709
|
brf1a
|
BRF1, RNA polymerase III transcription initiation factor a |
| chr11_+_37345219 | 0.55 |
ENSDART00000172899
|
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr6_-_11127409 | 0.55 |
ENSDART00000151125
|
pcnt
|
pericentrin |
| chr20_-_7079412 | 0.54 |
ENSDART00000040793
|
sirt5
|
sirtuin 5 |
| chr13_-_26668719 | 0.54 |
ENSDART00000139264
|
vrk2
|
vaccinia related kinase 2 |
| chr20_-_3301951 | 0.53 |
ENSDART00000123096
|
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr14_+_14536088 | 0.53 |
ENSDART00000158291
|
slbp
|
stem-loop binding protein |
| chr24_-_21199008 | 0.51 |
|
|
|
| chr12_-_5153412 | 0.51 |
ENSDART00000160037
|
fra10ac1
|
FRA10A associated CGG repeat 1 |
| chr24_-_19574761 | 0.50 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr24_-_31257458 | 0.50 |
ENSDART00000158808
|
cnn3a
|
calponin 3, acidic a |
| chr19_-_18689437 | 0.49 |
ENSDART00000016135
|
nfe2l3
|
nuclear factor, erythroid 2-like 3 |
| chr7_-_73629408 | 0.48 |
ENSDART00000124179
|
hist1h4l
|
histone 1, H4, like |
| chr1_+_45907424 | 0.48 |
ENSDART00000053240
|
cab39l
|
calcium binding protein 39-like |
| chr6_-_52677363 | 0.47 |
ENSDART00000083830
|
sdc4
|
syndecan 4 |
| chr21_+_34085267 | 0.45 |
ENSDART00000024750
|
hmgb3b
|
high mobility group box 3b |
| chr5_+_40235387 | 0.45 |
ENSDART00000147767
|
si:dkey-3h3.3
|
si:dkey-3h3.3 |
| chr22_+_17506147 | 0.44 |
ENSDART00000142871
|
hnrnpm
|
heterogeneous nuclear ribonucleoprotein M |
| KN149698v1_+_88937 | 0.43 |
ENSDART00000163341
|
ENSDARG00000100188
|
ENSDARG00000100188 |
| chr1_+_45267315 | 0.43 |
ENSDART00000108528
|
arhgef7b
|
Rho guanine nucleotide exchange factor (GEF) 7b |
| chr7_-_69883542 | 0.43 |
|
|
|
| chr7_-_69883581 | 0.43 |
|
|
|
| chr3_-_52406525 | 0.41 |
ENSDART00000138018
|
zgc:113210
|
zgc:113210 |
| chr22_-_10511906 | 0.41 |
ENSDART00000013933
|
si:dkey-42i9.4
|
si:dkey-42i9.4 |
| chr5_+_17508128 | 0.40 |
ENSDART00000048859
|
ascc2
|
activating signal cointegrator 1 complex subunit 2 |
| chr13_-_33348231 | 0.40 |
ENSDART00000160520
|
mad2l1bp
|
MAD2L1 binding protein |
| chr18_+_13235539 | 0.39 |
ENSDART00000032151
|
cotl1
|
coactosin-like F-actin binding protein 1 |
| chr13_+_42183685 | 0.39 |
ENSDART00000158367
|
ide
|
insulin-degrading enzyme |
| chr23_-_25871518 | 0.38 |
ENSDART00000041833
|
fitm2
|
fat storage-inducing transmembrane protein 2 |
| chr16_+_27608837 | 0.38 |
ENSDART00000147611
|
si:ch211-197h24.6
|
si:ch211-197h24.6 |
| chr13_+_42183490 | 0.38 |
ENSDART00000158367
|
ide
|
insulin-degrading enzyme |
| chr15_+_24629719 | 0.38 |
ENSDART00000134622
|
dhrs13b
|
dehydrogenase/reductase (SDR family) member 13b |
| chr9_+_33179226 | 0.37 |
|
|
|
| chr3_-_9504876 | 0.37 |
ENSDART00000128731
|
rnps1
|
RNA binding protein S1, serine-rich domain |
| chr19_-_23665476 | 0.37 |
ENSDART00000140665
|
grb10a
|
growth factor receptor-bound protein 10a |
| chr6_+_54568388 | 0.37 |
ENSDART00000093199
|
tead3b
|
TEA domain family member 3 b |
| chr2_+_30119672 | 0.35 |
ENSDART00000019417
|
dnajb6a
|
DnaJ (Hsp40) homolog, subfamily B, member 6a |
| chr25_+_25642475 | 0.35 |
ENSDART00000108476
|
lrrc61
|
leucine rich repeat containing 61 |
| chr17_-_7377094 | 0.35 |
|
|
|
| chr6_+_54568649 | 0.35 |
ENSDART00000157142
|
tead3b
|
TEA domain family member 3 b |
| chr11_-_3313199 | 0.34 |
ENSDART00000002545
|
mcrs1
|
microspherule protein 1 |
| chr5_+_61678410 | 0.34 |
ENSDART00000074117
|
aspa
|
aspartoacylase |
| chr5_-_37784663 | 0.33 |
ENSDART00000051233
|
mink1
|
misshapen-like kinase 1 |
| chr2_+_49723402 | 0.33 |
ENSDART00000129967
|
sh3gl1a
|
SH3-domain GRB2-like 1a |
| chr6_-_7577736 | 0.33 |
ENSDART00000151545
|
slc25a38b
|
solute carrier family 25, member 38b |
| chr16_+_25063153 | 0.33 |
ENSDART00000156579
|
si:dkeyp-84f3.5
|
si:dkeyp-84f3.5 |
| chr1_-_19375135 | 0.32 |
ENSDART00000102993
|
mettl14
|
methyltransferase like 14 |
| chr7_-_20378231 | 0.32 |
|
|
|
| chr18_-_20605032 | 0.31 |
ENSDART00000134722
|
bcl2l13
|
BCL2-like 13 (apoptosis facilitator) |
| chr23_-_34018069 | 0.31 |
ENSDART00000133223
|
ENSDARG00000070393
|
ENSDARG00000070393 |
| chr2_+_26584862 | 0.30 |
ENSDART00000129895
|
palm1b
|
paralemmin 1b |
| chr20_+_35156812 | 0.30 |
|
|
|
| chr6_+_54350379 | 0.28 |
ENSDART00000153704
|
anks1ab
|
ankyrin repeat and sterile alpha motif domain containing 1Ab |
| chr7_-_39874079 | 0.28 |
ENSDART00000173634
|
wdr60
|
WD repeat domain 60 |
| chr3_+_26113651 | 0.28 |
ENSDART00000103734
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr13_+_35402382 | 0.25 |
ENSDART00000163368
|
wdr27
|
WD repeat domain 27 |
| chr12_+_19199002 | 0.25 |
ENSDART00000066389
|
tmem184ba
|
transmembrane protein 184ba |
| chr11_+_26366532 | 0.25 |
ENSDART00000159505
|
dynlrb1
|
dynein, light chain, roadblock-type 1 |
| chr7_-_32624437 | 0.24 |
ENSDART00000141828
|
ano5b
|
anoctamin 5b |
| chr7_-_20378183 | 0.24 |
|
|
|
| chr24_-_24306432 | 0.24 |
ENSDART00000154149
|
BX323067.1
|
ENSDARG00000097984 |
| chr24_+_16402587 | 0.23 |
ENSDART00000164319
|
sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr1_+_16683891 | 0.23 |
ENSDART00000103262
ENSDART00000145068 ENSDART00000169619 ENSDART00000010526 |
fat1a
|
FAT atypical cadherin 1a |
| chr24_-_2453890 | 0.22 |
ENSDART00000093331
|
rreb1a
|
ras responsive element binding protein 1a |
| chr19_-_27755498 | 0.22 |
ENSDART00000137346
|
znrd1
|
zinc ribbon domain containing 1 |
| chr21_-_35806609 | 0.21 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr16_-_27608366 | 0.21 |
ENSDART00000147737
|
tex10
|
testis expressed 10 |
| chr13_+_15802118 | 0.21 |
ENSDART00000131390
|
fignl1
|
fidgetin-like 1 |
| chr16_+_33167509 | 0.21 |
ENSDART00000058472
|
akirin1
|
akirin 1 |
| chr10_-_26202233 | 0.21 |
ENSDART00000136472
|
trim3b
|
tripartite motif containing 3b |
| chr21_-_36711558 | 0.20 |
ENSDART00000086060
|
mrpl22
|
mitochondrial ribosomal protein L22 |
| chr1_+_35217331 | 0.20 |
|
|
|
| chr9_-_50304152 | 0.20 |
ENSDART00000161648
ENSDART00000168514 |
scn1a
|
sodium channel, voltage-gated, type I, alpha |
| chr16_-_36061564 | 0.19 |
ENSDART00000175847
|
CABZ01065723.1
|
ENSDARG00000105999 |
| chr11_-_42934097 | 0.19 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr16_-_22394669 | 0.18 |
|
|
|
| chr3_+_26113393 | 0.18 |
ENSDART00000103733
|
atad5a
|
ATPase family, AAA domain containing 5a |
| chr10_-_28142344 | 0.17 |
ENSDART00000023545
|
ints2
|
integrator complex subunit 2 |
| chr7_+_22530248 | 0.17 |
ENSDART00000148054
|
rbm4.1
|
RNA binding motif protein 4.1 |
| chr25_+_25642342 | 0.16 |
ENSDART00000108476
|
lrrc61
|
leucine rich repeat containing 61 |
| chr19_+_7505348 | 0.15 |
ENSDART00000004622
|
sf3b4
|
splicing factor 3b, subunit 4 |
| chr16_-_33167598 | 0.15 |
|
|
|
| chr7_+_26977165 | 0.15 |
|
|
|
| chr24_-_40968527 | 0.15 |
ENSDART00000172617
|
smyhc1
|
slow myosin heavy chain 1 |
| chr25_+_2515462 | 0.14 |
ENSDART00000092919
ENSDART00000122929 |
bbs4
|
Bardet-Biedl syndrome 4 |
| chr2_-_57020554 | 0.14 |
|
|
|
| chr15_+_12497915 | 0.14 |
ENSDART00000168997
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr15_-_11798324 | 0.13 |
|
|
|
| chr3_+_62051743 | 0.13 |
ENSDART00000168063
|
zgc:173575
|
zgc:173575 |
| chr15_-_5911297 | 0.12 |
|
|
|
| chr7_-_6323664 | 0.12 |
ENSDART00000173158
|
zgc:112234
|
zgc:112234 |
| chr5_-_11405633 | 0.11 |
ENSDART00000114378
|
fbxw8
|
F-box and WD repeat domain containing 8 |
| chr15_-_5911817 | 0.11 |
|
|
|
| chr7_+_73603232 | 0.10 |
ENSDART00000112685
|
hist1h4l
|
histone 1, H4, like |
| chr5_+_30880165 | 0.10 |
ENSDART00000098197
|
ENSDARG00000035471
|
ENSDARG00000035471 |
| chr24_+_36504391 | 0.09 |
ENSDART00000135898
|
arf2b
|
ADP-ribosylation factor 2b |
| chr1_+_39276921 | 0.09 |
ENSDART00000166251
|
aip
|
aryl hydrocarbon receptor interacting protein |
| chr19_+_22763741 | 0.09 |
ENSDART00000109157
|
fbxl6
|
F-box and leucine-rich repeat protein 6 |
| chr19_-_44043912 | 0.08 |
ENSDART00000138009
ENSDART00000086138 |
fam83hb
|
family with sequence similarity 83, member Hb |
| chr25_+_25642290 | 0.08 |
ENSDART00000108476
|
lrrc61
|
leucine rich repeat containing 61 |
| chr25_-_22791921 | 0.08 |
ENSDART00000128250
|
mob2a
|
MOB kinase activator 2a |
| chr20_+_7596461 | 0.08 |
ENSDART00000127975
ENSDART00000132481 ENSDART00000144551 |
bloc1s2
|
biogenesis of lysosomal organelles complex-1, subunit 2 |
| chr24_-_24578984 | 0.07 |
ENSDART00000012399
|
armc1
|
armadillo repeat containing 1 |
| chr25_+_34357376 | 0.07 |
ENSDART00000149782
|
CHST6
|
carbohydrate sulfotransferase 6 |
| chr1_-_10217300 | 0.07 |
|
|
|
| chr7_-_13109764 | 0.06 |
ENSDART00000091616
|
sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr7_+_23735703 | 0.06 |
ENSDART00000033755
|
homezb
|
homeobox and leucine zipper encoding b |
| chr14_-_5509554 | 0.06 |
|
|
|
| chr14_+_6655541 | 0.06 |
ENSDART00000148447
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr24_-_34794463 | 0.06 |
ENSDART00000171009
ENSDART00000170046 |
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr2_+_47769055 | 0.05 |
ENSDART00000154465
|
si:ch211-165b10.3
|
si:ch211-165b10.3 |
| chr19_-_44043954 | 0.05 |
ENSDART00000138009
ENSDART00000086138 |
fam83hb
|
family with sequence similarity 83, member Hb |
| chr9_-_30752583 | 0.05 |
|
|
|
| chr7_-_24724452 | 0.04 |
ENSDART00000173791
|
rcor2
|
REST corepressor 2 |
| chr24_-_25189211 | 0.04 |
|
|
|
| chr22_-_10540917 | 0.04 |
ENSDART00000140736
|
si:dkey-42i9.6
|
si:dkey-42i9.6 |
| chr15_+_32439470 | 0.04 |
ENSDART00000153657
|
trim3a
|
tripartite motif containing 3a |
| chr12_-_30894479 | 0.04 |
ENSDART00000133854
|
tcf7l2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr20_-_20455731 | 0.04 |
ENSDART00000170488
ENSDART00000047580 |
hif1ab
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) b |
| chr5_+_21661082 | 0.03 |
|
|
|
| chr9_-_7419306 | 0.03 |
ENSDART00000125054
|
slc23a3
|
solute carrier family 23, member 3 |
| chr15_-_5911548 | 0.03 |
|
|
|
| KN150339v1_-_34277 | 0.02 |
|
|
|
| chr11_+_31061296 | 0.02 |
ENSDART00000088093
|
sipa1l2
|
signal-induced proliferation-associated 1 like 2 |
| chr14_-_2379419 | 0.01 |
|
|
|
| chr14_+_6656015 | 0.00 |
ENSDART00000148394
|
hnrnpaba
|
heterogeneous nuclear ribonucleoprotein A/Ba |
| chr6_-_34950360 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.2 | 0.8 | GO:0010482 | keratinocyte development(GO:0003334) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 1.0 | GO:1904666 | regulation of ubiquitin protein ligase activity(GO:1904666) |
| 0.1 | 0.4 | GO:2000815 | regulation of mRNA stability involved in response to stress(GO:0010610) regulation of mRNA stability involved in response to oxidative stress(GO:2000815) |
| 0.1 | 0.7 | GO:0003379 | establishment of cell polarity involved in gastrulation cell migration(GO:0003379) |
| 0.1 | 0.5 | GO:0036046 | regulation of ketone biosynthetic process(GO:0010566) protein demalonylation(GO:0036046) peptidyl-lysine demalonylation(GO:0036047) protein desuccinylation(GO:0036048) peptidyl-lysine desuccinylation(GO:0036049) protein deglutarylation(GO:0061698) |
| 0.1 | 1.0 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.1 | 0.6 | GO:0036149 | phosphatidylinositol acyl-chain remodeling(GO:0036149) |
| 0.1 | 1.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.4 | GO:0048103 | neuronal stem cell division(GO:0036445) somatic stem cell division(GO:0048103) |
| 0.1 | 0.4 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.1 | 0.3 | GO:0021861 | forebrain radial glial cell differentiation(GO:0021861) |
| 0.1 | 1.0 | GO:0032196 | transposition(GO:0032196) |
| 0.1 | 2.9 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.1 | 0.8 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.1 | 0.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.1 | 1.2 | GO:0035914 | skeletal muscle cell differentiation(GO:0035914) |
| 0.1 | 0.2 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) regulation of macrophage chemotaxis(GO:0010758) positive regulation of macrophage chemotaxis(GO:0010759) skeletal muscle satellite cell proliferation(GO:0014841) regulation of skeletal muscle satellite cell proliferation(GO:0014842) negative regulation of skeletal muscle cell proliferation(GO:0014859) mononuclear cell migration(GO:0071674) regulation of mononuclear cell migration(GO:0071675) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 1.1 | GO:0030223 | neutrophil differentiation(GO:0030223) DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.4 | GO:0034389 | coenzyme catabolic process(GO:0009109) lipid particle organization(GO:0034389) |
| 0.0 | 1.3 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.4 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 2.3 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.4 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.4 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.6 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.7 | GO:0060059 | positive regulation of BMP signaling pathway(GO:0030513) embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.1 | GO:0097032 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.9 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 1.8 | GO:0016042 | lipid catabolic process(GO:0016042) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.2 | 0.8 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.4 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 0.6 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.3 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.6 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.4 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.9 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.3 | GO:0044545 | NSL complex(GO:0044545) |
| 0.0 | 1.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.9 | 2.6 | GO:0017125 | deoxycytidyl transferase activity(GO:0017125) |
| 0.3 | 1.1 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 0.5 | GO:0036054 | protein-malonyllysine demalonylase activity(GO:0036054) protein-succinyllysine desuccinylase activity(GO:0036055) protein-glutaryllysine deglutarylase activity(GO:0061697) |
| 0.1 | 0.3 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.3 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.1 | 1.0 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 1.4 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.1 | 0.7 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.3 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.6 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.6 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.0 | 0.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 2.0 | GO:0016811 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides(GO:0016811) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.4 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.5 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.4 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.3 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.6 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.7 | GO:0017147 | Wnt-protein binding(GO:0017147) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 2.9 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 1.0 | PID PLK1 PATHWAY | PLK1 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 0.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.5 | REACTOME ENERGY DEPENDENT REGULATION OF MTOR BY LKB1 AMPK | Genes involved in Energy dependent regulation of mTOR by LKB1-AMPK |
| 0.0 | 1.3 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 1.4 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 2.6 | REACTOME DNA REPAIR | Genes involved in DNA Repair |
| 0.0 | 0.4 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.6 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.5 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |