Project
DANIO-CODE
Navigation
Downloads
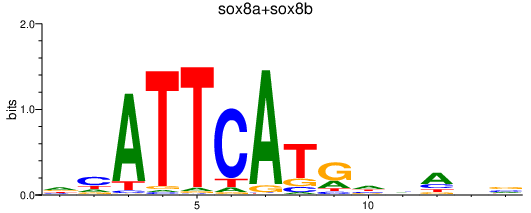
Results for sox8a+sox8b
Z-value: 0.38
Transcription factors associated with sox8a+sox8b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox8b
|
ENSDARG00000037782 | SRY-box transcription factor 8b |
|
sox8a
|
ENSDARG00000105301 | SRY-box transcription factor 8a |
Activity profile of sox8a+sox8b motif
Sorted Z-values of sox8a+sox8b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of sox8a+sox8b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_12484651 | 0.73 |
ENSDART00000162258
|
BX901957.1
|
ENSDARG00000098363 |
| chr17_+_2373108 | 0.72 |
ENSDART00000156843
|
si:dkey-248g15.3
|
si:dkey-248g15.3 |
| chr24_-_21778717 | 0.58 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr3_-_27516974 | 0.55 |
ENSDART00000151675
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr23_-_35595271 | 0.51 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr14_+_38445969 | 0.50 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr5_-_6004819 | 0.46 |
ENSDART00000099570
|
tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr15_+_28435937 | 0.45 |
ENSDART00000142298
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr25_+_24520476 | 0.42 |
|
|
|
| chr2_-_24898678 | 0.41 |
ENSDART00000145145
|
cnn2
|
calponin 2 |
| chr13_+_33151628 | 0.40 |
ENSDART00000135200
|
ccdc28b
|
coiled-coil domain containing 28B |
| chr11_-_18027225 | 0.39 |
|
|
|
| chr17_-_32913432 | 0.37 |
ENSDART00000077476
|
prox1a
|
prospero homeobox 1a |
| chr3_-_6922655 | 0.36 |
|
|
|
| chr2_+_10148323 | 0.33 |
ENSDART00000158214
|
tsen15
|
TSEN15 tRNA splicing endonuclease subunit |
| chr3_-_27517052 | 0.32 |
ENSDART00000151625
|
si:ch211-157c3.4
|
si:ch211-157c3.4 |
| chr24_-_41356844 | 0.30 |
ENSDART00000121592
ENSDART00000019975 ENSDART00000166034 |
acvr2b
|
activin A receptor, type IIB |
| chr21_+_43674754 | 0.30 |
ENSDART00000136025
|
tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr14_+_6239755 | 0.28 |
ENSDART00000097214
|
adam19b
|
ADAM metallopeptidase domain 19b |
| chr11_-_37730181 | 0.28 |
ENSDART00000102870
|
slc41a1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr5_+_69036145 | 0.26 |
ENSDART00000174403
|
fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr9_-_43736549 | 0.25 |
ENSDART00000140526
|
znf385b
|
zinc finger protein 385B |
| chr13_-_8560564 | 0.25 |
ENSDART00000144553
|
plekhh2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| chr7_-_50137683 | 0.24 |
ENSDART00000083346
|
hypk
|
huntingtin interacting protein K |
| chr7_+_64897785 | 0.22 |
ENSDART00000113602
ENSDART00000167822 |
ipo7
|
importin 7 |
| chr11_-_29891067 | 0.22 |
ENSDART00000172106
|
scml2
|
sex comb on midleg-like 2 (Drosophila) |
| chr19_-_27864181 | 0.22 |
ENSDART00000079251
|
vars2
|
valyl-tRNA synthetase 2, mitochondrial (putative) |
| chr20_+_54525614 | 0.21 |
ENSDART00000160409
|
arf6a
|
ADP-ribosylation factor 6a |
| chr17_+_24668907 | 0.21 |
ENSDART00000034263
ENSDART00000135794 |
sepn1
|
selenoprotein N, 1 |
| chr7_-_18295101 | 0.20 |
ENSDART00000173969
|
rgs12a
|
regulator of G protein signaling 12a |
| chr6_+_7165326 | 0.20 |
ENSDART00000065500
|
abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr3_+_41584507 | 0.20 |
ENSDART00000154401
|
chst12a
|
carbohydrate (chondroitin 4) sulfotransferase 12a |
| chr22_-_38635149 | 0.20 |
ENSDART00000098330
|
nppc
|
natriuretic peptide C |
| chr2_+_10148394 | 0.19 |
ENSDART00000158214
|
tsen15
|
TSEN15 tRNA splicing endonuclease subunit |
| chr4_+_22576862 | 0.19 |
ENSDART00000077707
|
llph
|
LLP homolog, long-term synaptic facilitation (Aplysia) |
| chr7_-_52574822 | 0.18 |
ENSDART00000172951
|
map1aa
|
microtubule-associated protein 1Aa |
| chr8_+_46378250 | 0.17 |
ENSDART00000129661
ENSDART00000084081 |
ogg1
|
8-oxoguanine DNA glycosylase |
| chr1_-_9256864 | 0.17 |
ENSDART00000041849
|
tmem8a
|
transmembrane protein 8A |
| chr16_-_25691856 | 0.17 |
ENSDART00000077447
ENSDART00000131528 |
zgc:110410
|
zgc:110410 |
| chr20_-_54669179 | 0.16 |
|
|
|
| chr1_+_5948527 | 0.16 |
ENSDART00000138919
|
ube2f
|
ubiquitin-conjugating enzyme E2F (putative) |
| chr16_+_25180444 | 0.15 |
ENSDART00000157346
ENSDART00000155032 |
si:ch211-261d7.3
|
si:ch211-261d7.3 |
| chr7_-_38521262 | 0.15 |
ENSDART00000157416
|
si:dkey-23n7.10
|
si:dkey-23n7.10 |
| chr8_-_46378116 | 0.15 |
ENSDART00000136602
|
qars
|
glutaminyl-tRNA synthetase |
| chr1_+_37643126 | 0.14 |
ENSDART00000075086
|
cep44
|
centrosomal protein 44 |
| chr17_+_14776302 | 0.14 |
ENSDART00000154229
|
RTRAF
|
zgc:56576 |
| chr19_-_27284726 | 0.14 |
ENSDART00000089699
|
prrt1
|
proline-rich transmembrane protein 1 |
| chr10_+_16009487 | 0.14 |
ENSDART00000136709
|
apbb1
|
amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) |
| chr21_-_43332911 | 0.13 |
ENSDART00000114955
|
sowahaa
|
sosondowah ankyrin repeat domain family member Aa |
| chr6_-_37765778 | 0.13 |
ENSDART00000149068
|
nipa2
|
non imprinted in Prader-Willi/Angelman syndrome 2 (human) |
| chr14_-_25154966 | 0.12 |
ENSDART00000148652
|
slc26a2
|
solute carrier family 26 (anion exchanger), member 2 |
| chr10_+_9762039 | 0.12 |
|
|
|
| chr19_-_27864288 | 0.11 |
ENSDART00000079251
|
vars2
|
valyl-tRNA synthetase 2, mitochondrial (putative) |
| chr5_+_18797669 | 0.11 |
ENSDART00000165119
|
rusc2
|
RUN and SH3 domain containing 2 |
| chr6_+_13273286 | 0.10 |
ENSDART00000032331
|
gmppab
|
GDP-mannose pyrophosphorylase Ab |
| chr9_+_17415990 | 0.09 |
ENSDART00000112884
|
kbtbd7
|
kelch repeat and BTB (POZ) domain containing 7 |
| chr12_+_30591482 | 0.09 |
ENSDART00000133869
|
kcnk1a
|
potassium channel, subfamily K, member 1a |
| chr10_-_14587464 | 0.08 |
ENSDART00000057865
|
ier3ip1
|
immediate early response 3 interacting protein 1 |
| chr21_+_37668334 | 0.07 |
ENSDART00000143621
|
nsd1b
|
nuclear receptor binding SET domain protein 1b |
| chr18_-_20880226 | 0.07 |
ENSDART00000090079
|
synm
|
synemin, intermediate filament protein |
| chr8_+_20592099 | 0.07 |
ENSDART00000160421
|
nfic
|
nuclear factor I/C |
| chr9_-_3028101 | 0.07 |
ENSDART00000144046
|
rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr21_-_22441376 | 0.06 |
|
|
|
| chr13_-_8560620 | 0.06 |
ENSDART00000144553
|
plekhh2
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 2 |
| chr2_+_25114775 | 0.06 |
ENSDART00000078854
|
mpv17l2
|
MPV17 mitochondrial membrane protein-like 2 |
| chr17_+_11961595 | 0.06 |
|
|
|
| chr5_+_54642472 | 0.05 |
ENSDART00000131060
|
pcsk5a
|
proprotein convertase subtilisin/kexin type 5a |
| chr15_+_28435875 | 0.04 |
ENSDART00000168453
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr14_+_49875056 | 0.04 |
ENSDART00000167810
|
zgc:154054
|
zgc:154054 |
| chr7_+_22710714 | 0.04 |
ENSDART00000122449
|
ccnb3
|
cyclin B3 |
| chr20_+_30676086 | 0.03 |
ENSDART00000131422
ENSDART00000010494 |
fgfr1op
|
FGFR1 oncogene partner |
| chr6_+_49927433 | 0.03 |
ENSDART00000018523
|
ahcy
|
adenosylhomocysteinase |
| chr2_+_42923652 | 0.03 |
ENSDART00000168318
|
BX005083.1
|
ENSDARG00000097562 |
| chr4_+_6825027 | 0.02 |
ENSDART00000165179
|
dock4b
|
dedicator of cytokinesis 4b |
| chr19_-_12032026 | 0.02 |
ENSDART00000024861
ENSDART00000110513 ENSDART00000163478 |
cpsf1
|
cleavage and polyadenylation specific factor 1 |
| chr20_+_26037296 | 0.01 |
ENSDART00000141265
|
znf106b
|
zinc finger protein 106b |
| chr11_+_39694880 | 0.01 |
ENSDART00000024304
|
per3
|
period circadian clock 3 |
| chr10_+_5267746 | 0.01 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr20_+_31371853 | 0.01 |
|
|
|
| chr20_-_29602821 | 0.01 |
ENSDART00000148278
|
klf11a
|
Kruppel-like factor 11a |
| chr8_-_14014576 | 0.00 |
ENSDART00000135811
|
atp2b3a
|
ATPase, Ca++ transporting, plasma membrane 3a |
| chr16_+_50355723 | 0.00 |
ENSDART00000166664
|
ENSDARG00000102916
|
ENSDARG00000102916 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.0 | 0.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.3 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.0 | 0.5 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.4 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:0036010 | protein localization to endosome(GO:0036010) |
| 0.0 | 0.4 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.5 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.0 | 0.1 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.2 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.3 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.3 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.2 | GO:0031114 | regulation of microtubule depolymerization(GO:0031114) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.3 | GO:0002161 | aminoacyl-tRNA editing activity(GO:0002161) |
| 0.0 | 0.1 | GO:0008905 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 0.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 0.1 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.5 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.2 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |