Project
DANIO-CODE
Navigation
Downloads
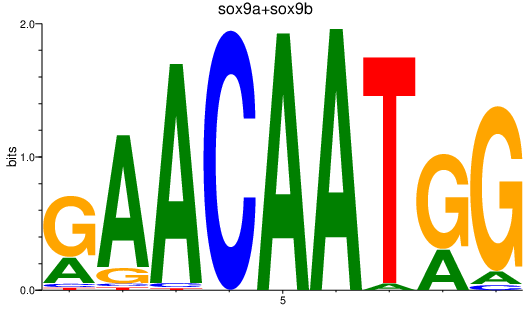
Results for sox9a+sox9b
Z-value: 1.36
Transcription factors associated with sox9a+sox9b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sox9a
|
ENSDARG00000003293 | SRY-box transcription factor 9a |
|
sox9b
|
ENSDARG00000043923 | SRY-box transcription factor 9b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sox9a | dr10_dc_chr12_-_1931281_1931287 | 0.82 | 8.4e-05 | Click! |
| sox9b | dr10_dc_chr3_-_62348450_62348451 | 0.19 | 4.7e-01 | Click! |
Activity profile of sox9a+sox9b motif
Sorted Z-values of sox9a+sox9b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of sox9a+sox9b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_12733209 | 5.94 |
ENSDART00000167021
|
mcm6
|
minichromosome maintenance complex component 6 |
| chr14_+_9115745 | 4.06 |
ENSDART00000123652
|
hmgn6
|
high mobility group nucleosome binding domain 6 |
| chr23_+_35609887 | 3.44 |
ENSDART00000179393
|
tuba1b
|
tubulin, alpha 1b |
| chr5_+_34857986 | 3.42 |
ENSDART00000141239
|
erlin2
|
ER lipid raft associated 2 |
| chr16_+_23482744 | 3.19 |
ENSDART00000148961
|
s100a10b
|
S100 calcium binding protein A10b |
| chr15_+_29153215 | 3.13 |
ENSDART00000156799
|
si:ch211-137a8.4
|
si:ch211-137a8.4 |
| chr3_-_35928594 | 2.42 |
|
|
|
| chr14_+_38445969 | 2.34 |
ENSDART00000164440
|
si:ch211-195b11.3
|
si:ch211-195b11.3 |
| chr13_+_28601627 | 2.15 |
ENSDART00000015773
|
ldb1a
|
LIM domain binding 1a |
| chr4_+_57435 | 2.12 |
ENSDART00000169187
|
ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr6_+_29800606 | 2.12 |
ENSDART00000017424
|
ptmaa
|
prothymosin, alpha a |
| chr24_-_24306469 | 2.07 |
ENSDART00000154149
|
BX323067.1
|
ENSDARG00000097984 |
| chr13_+_1051015 | 2.03 |
ENSDART00000033528
|
tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr11_-_41357639 | 2.02 |
ENSDART00000055709
|
her2
|
hairy-related 2 |
| chr14_+_11151485 | 2.01 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr11_-_29910947 | 2.01 |
ENSDART00000156121
|
scml2
|
sex comb on midleg-like 2 (Drosophila) |
| chr23_+_24711233 | 1.98 |
|
|
|
| chr6_-_43094573 | 1.98 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr24_+_9335428 | 1.96 |
ENSDART00000132688
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr3_-_16100715 | 1.94 |
ENSDART00000146699
|
bckdhbl
|
branched chain keto acid dehydrogenase E1, beta polypeptide, like |
| chr9_-_54126121 | 1.93 |
ENSDART00000126314
|
pcdh8
|
protocadherin 8 |
| chr6_+_47845002 | 1.90 |
ENSDART00000140943
|
padi2
|
peptidyl arginine deiminase, type II |
| chr4_-_8610868 | 1.89 |
ENSDART00000067322
|
fbxl14b
|
F-box and leucine-rich repeat protein 14b |
| chr17_-_36988937 | 1.86 |
ENSDART00000145236
|
dpysl5a
|
dihydropyrimidinase-like 5a |
| chr19_-_3226831 | 1.83 |
ENSDART00000145710
ENSDART00000110763 ENSDART00000074620 |
stm
|
starmaker |
| chr14_-_26406720 | 1.82 |
ENSDART00000078563
|
neurog1
|
neurogenin 1 |
| chr16_+_16941228 | 1.81 |
ENSDART00000142155
|
myh14
|
myosin, heavy chain 14, non-muscle |
| chr23_-_30119058 | 1.80 |
ENSDART00000103480
|
ccdc187
|
coiled-coil domain containing 187 |
| chr3_+_28450576 | 1.78 |
ENSDART00000150893
|
sept12
|
septin 12 |
| chr5_-_51109419 | 1.78 |
ENSDART00000163464
|
lhfpl2b
|
lipoma HMGIC fusion partner-like 2b |
| chr17_+_34854673 | 1.78 |
|
|
|
| chr24_+_9335316 | 1.75 |
ENSDART00000132688
|
si:ch211-285f17.1
|
si:ch211-285f17.1 |
| chr14_+_23980348 | 1.74 |
ENSDART00000160984
|
BX005228.1
|
ENSDARG00000105465 |
| chr19_-_18316262 | 1.74 |
ENSDART00000151133
|
top2b
|
topoisomerase (DNA) II beta |
| chr8_+_23071884 | 1.74 |
ENSDART00000063075
|
zgc:100920
|
zgc:100920 |
| chr20_+_6640497 | 1.74 |
ENSDART00000138361
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr3_+_56970554 | 1.71 |
ENSDART00000162930
|
bahcc1a
|
BAH domain and coiled-coil containing 1a |
| chr22_-_21021645 | 1.70 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr22_-_37417903 | 1.67 |
ENSDART00000149948
|
FP102167.1
|
ENSDARG00000095844 |
| chr25_+_30699938 | 1.67 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr20_-_20412830 | 1.65 |
ENSDART00000114779
|
ENSDARG00000079369
|
ENSDARG00000079369 |
| chr3_+_61924544 | 1.63 |
ENSDART00000090370
|
noxo1a
|
NADPH oxidase organizer 1a |
| chr11_-_29316162 | 1.62 |
ENSDART00000163958
|
arhgef10la
|
Rho guanine nucleotide exchange factor (GEF) 10-like a |
| chr21_-_5852663 | 1.60 |
ENSDART00000130521
|
CACFD1
|
calcium channel flower domain containing 1 |
| chr25_-_13737344 | 1.56 |
|
|
|
| chr22_+_20436287 | 1.56 |
ENSDART00000135984
|
onecut3a
|
one cut homeobox 3a |
| chr10_-_10372266 | 1.55 |
|
|
|
| chr21_-_37509454 | 1.51 |
ENSDART00000175126
|
fgfr4
|
fibroblast growth factor receptor 4 |
| chr15_-_18110169 | 1.51 |
|
|
|
| chr3_-_58488929 | 1.51 |
ENSDART00000042386
|
unm_sa1261
|
un-named sa1261 |
| chr3_-_40263048 | 1.49 |
ENSDART00000154236
|
AL928657.1
|
ENSDARG00000097119 |
| chr19_+_44373777 | 1.49 |
ENSDART00000051712
|
gatad1
|
GATA zinc finger domain containing 1 |
| chr15_-_39756101 | 1.48 |
|
|
|
| chr10_-_11053655 | 1.47 |
ENSDART00000132995
|
ak3
|
adenylate kinase 3 |
| chr14_+_34683602 | 1.47 |
ENSDART00000172171
|
ebf1a
|
early B-cell factor 1a |
| chr13_-_22901327 | 1.45 |
ENSDART00000056523
|
hkdc1
|
hexokinase domain containing 1 |
| chr1_-_46141359 | 1.44 |
ENSDART00000142406
|
si:ch73-160h15.3
|
si:ch73-160h15.3 |
| chr21_+_25651712 | 1.44 |
ENSDART00000135123
|
si:dkey-17e16.17
|
si:dkey-17e16.17 |
| chr11_+_18823629 | 1.44 |
|
|
|
| chr3_+_32394653 | 1.44 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr3_+_33629940 | 1.44 |
ENSDART00000169337
|
ier2a
|
immediate early response 2a |
| chr15_-_4537178 | 1.43 |
ENSDART00000155619
ENSDART00000128602 |
tfdp2
|
transcription factor Dp-2 |
| chr25_-_22821132 | 1.40 |
|
|
|
| chr14_-_33637792 | 1.40 |
ENSDART00000128515
ENSDART00000125254 |
foxa
|
forkhead box A sequence |
| chr10_+_37139869 | 1.39 |
ENSDART00000132162
|
vezf1a
|
vascular endothelial zinc finger 1a |
| chr24_+_16998329 | 1.37 |
ENSDART00000177272
|
mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr1_-_34921848 | 1.35 |
ENSDART00000142154
|
frem3
|
Fras1 related extracellular matrix 3 |
| chr9_+_30279907 | 1.35 |
ENSDART00000102981
|
col8a1a
|
collagen, type VIII, alpha 1a |
| chr3_+_21059221 | 1.35 |
ENSDART00000078807
|
zgc:123295
|
zgc:123295 |
| chr19_+_47719438 | 1.34 |
ENSDART00000138295
|
ext1c
|
exostoses (multiple) 1c |
| chr17_+_16557246 | 1.34 |
ENSDART00000015729
ENSDART00000136874 |
foxn3
|
forkhead box N3 |
| chr8_+_25235278 | 1.34 |
ENSDART00000143554
|
ampd2b
|
adenosine monophosphate deaminase 2b |
| chr23_+_38239701 | 1.29 |
ENSDART00000148188
|
zgc:112994
|
zgc:112994 |
| chr6_-_12616971 | 1.28 |
ENSDART00000056764
|
bmpr2a
|
bone morphogenetic protein receptor, type II a (serine/threonine kinase) |
| chr4_-_5325164 | 1.28 |
ENSDART00000067375
|
osgep
|
O-sialoglycoprotein endopeptidase |
| chr17_-_33463482 | 1.27 |
ENSDART00000164064
|
ccdc28a
|
coiled-coil domain containing 28A |
| chr2_+_3125365 | 1.27 |
ENSDART00000164308
ENSDART00000167649 |
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr5_-_33006537 | 1.26 |
ENSDART00000048350
|
si:dkey-34e4.1
|
si:dkey-34e4.1 |
| chr12_-_22438379 | 1.26 |
ENSDART00000177715
|
CR925753.1
|
ENSDARG00000108493 |
| chr19_-_2079482 | 1.26 |
ENSDART00000128639
|
ENSDARG00000086326
|
ENSDARG00000086326 |
| chr12_+_27444832 | 1.25 |
ENSDART00000133719
|
etv4
|
ets variant 4 |
| chr9_-_22528568 | 1.25 |
ENSDART00000134805
|
crygm2d1
|
crystallin, gamma M2d1 |
| chr5_+_58727722 | 1.25 |
ENSDART00000082983
|
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr24_+_38783264 | 1.24 |
ENSDART00000154214
|
si:ch73-70c5.1
|
si:ch73-70c5.1 |
| chr23_-_18972097 | 1.23 |
ENSDART00000133826
|
CR847953.1
|
ENSDARG00000057403 |
| chr2_+_38178934 | 1.22 |
ENSDART00000044264
|
mmp14b
|
matrix metallopeptidase 14b (membrane-inserted) |
| chr9_-_2588401 | 1.22 |
ENSDART00000161018
|
sp9
|
sp9 transcription factor |
| chr23_-_30858769 | 1.21 |
ENSDART00000131285
|
myt1a
|
myelin transcription factor 1a |
| chr4_+_14972708 | 1.21 |
ENSDART00000005985
|
smo
|
smoothened, frizzled class receptor |
| chr21_-_35806638 | 1.20 |
ENSDART00000172245
|
sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr23_-_31446156 | 1.19 |
ENSDART00000053367
|
hmgn3
|
high mobility group nucleosomal binding domain 3 |
| chr22_+_15480482 | 1.19 |
ENSDART00000125450
|
gpc1a
|
glypican 1a |
| chr15_+_23615727 | 1.18 |
ENSDART00000152720
|
MARK4
|
microtubule affinity regulating kinase 4 |
| chr19_-_41887040 | 1.17 |
ENSDART00000062026
|
dlx5a
|
distal-less homeobox 5a |
| chr10_+_38582701 | 1.17 |
ENSDART00000144329
|
acer3
|
alkaline ceramidase 3 |
| chr5_+_44316830 | 1.16 |
|
|
|
| chr7_-_57796486 | 1.15 |
ENSDART00000043984
|
ank2b
|
ankyrin 2b, neuronal |
| chr7_+_16610457 | 1.15 |
|
|
|
| chr15_+_19717045 | 1.14 |
ENSDART00000138680
|
lim2.3
|
lens intrinsic membrane protein 2.3 |
| chr14_-_2079442 | 1.13 |
|
|
|
| chr3_+_32557992 | 1.12 |
ENSDART00000029262
|
si:dkey-16l2.17
|
si:dkey-16l2.17 |
| chr25_+_19992389 | 1.12 |
ENSDART00000143441
|
tnni4b.2
|
troponin I4b, tandem duplicate 2 |
| chr19_+_5399813 | 1.12 |
ENSDART00000145749
|
si:dkeyp-113d7.1
|
si:dkeyp-113d7.1 |
| chr11_+_24687813 | 1.11 |
ENSDART00000131431
|
sulf2a
|
sulfatase 2a |
| chr10_+_18994733 | 1.10 |
ENSDART00000146517
|
dpysl2b
|
dihydropyrimidinase-like 2b |
| chr5_-_31763197 | 1.10 |
ENSDART00000098045
|
gas1b
|
growth arrest-specific 1b |
| chr4_-_12719490 | 1.09 |
ENSDART00000035259
|
mgst1.1
|
microsomal glutathione S-transferase 1.1 |
| chr1_+_46824595 | 1.08 |
|
|
|
| chr24_+_24308055 | 1.07 |
|
|
|
| chr9_-_10561062 | 1.07 |
ENSDART00000175269
|
thsd7ba
|
thrombospondin, type I, domain containing 7Ba |
| chr2_+_11902170 | 1.07 |
ENSDART00000138562
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr20_-_40553998 | 1.06 |
ENSDART00000075092
|
pkib
|
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chr6_-_43094926 | 1.06 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr9_-_31467299 | 1.06 |
ENSDART00000022204
|
zic5
|
zic family member 5 (odd-paired homolog, Drosophila) |
| chr25_-_36760720 | 1.05 |
ENSDART00000111862
|
ldlrad3
|
low density lipoprotein receptor class A domain containing 3 |
| chr13_-_508202 | 1.05 |
ENSDART00000066080
ENSDART00000128969 |
slit1a
|
slit homolog 1a (Drosophila) |
| chr20_-_3302116 | 1.04 |
ENSDART00000123096
|
marcksa
|
myristoylated alanine-rich protein kinase C substrate a |
| chr3_+_52745202 | 1.04 |
ENSDART00000104683
|
pbx4
|
pre-B-cell leukemia transcription factor 4 |
| chr2_+_56339323 | 1.04 |
ENSDART00000123392
|
rab11bb
|
RAB11B, member RAS oncogene family, b |
| chr19_-_18316039 | 1.03 |
ENSDART00000151133
|
top2b
|
topoisomerase (DNA) II beta |
| chr25_-_31881585 | 1.01 |
ENSDART00000123999
ENSDART00000158091 |
ENSDARG00000100731
|
ENSDARG00000100731 |
| chr16_-_6881218 | 1.01 |
ENSDART00000149070
|
mbpb
|
myelin basic protein b |
| chr1_+_46537108 | 1.01 |
|
|
|
| chr17_+_43495818 | 1.00 |
ENSDART00000149041
|
reep1
|
receptor accessory protein 1 |
| chr14_-_2044362 | 1.00 |
|
|
|
| chr23_-_33728344 | 1.00 |
ENSDART00000146180
|
csrnp2
|
cysteine-serine-rich nuclear protein 2 |
| chr5_-_36349284 | 1.00 |
ENSDART00000047269
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr10_-_31838886 | 0.98 |
ENSDART00000128839
|
fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr4_+_28429523 | 0.98 |
ENSDART00000111603
|
znf1041
|
zinc finger protein 1041 |
| chr6_-_24003717 | 0.98 |
ENSDART00000164915
|
scinla
|
scinderin like a |
| chr21_-_9708608 | 0.96 |
|
|
|
| chr10_-_1690605 | 0.96 |
ENSDART00000137620
|
si:ch73-46j18.5
|
si:ch73-46j18.5 |
| chr18_-_40865967 | 0.96 |
ENSDART00000098865
|
vaspb
|
vasodilator-stimulated phosphoprotein b |
| chr12_+_16232466 | 0.95 |
ENSDART00000152500
|
ppp1r3cb
|
protein phosphatase 1, regulatory subunit 3Cb |
| chr7_-_51046010 | 0.95 |
ENSDART00000067647
|
rasl11a
|
RAS-like, family 11, member A |
| chr17_-_37107310 | 0.95 |
ENSDART00000151921
|
dnmt3ab
|
DNA (cytosine-5-)-methyltransferase 3 alpha b |
| chr6_+_12618821 | 0.94 |
ENSDART00000156341
|
fam117ba
|
family with sequence similarity 117, member Ba |
| chr1_-_10391257 | 0.94 |
ENSDART00000102903
|
dmd
|
dystrophin |
| chr15_-_17933972 | 0.93 |
ENSDART00000155066
|
atf5b
|
activating transcription factor 5b |
| chr19_+_38033219 | 0.92 |
ENSDART00000158960
|
thsd7aa
|
thrombospondin, type I, domain containing 7Aa |
| chr6_+_2849460 | 0.92 |
|
|
|
| chr17_-_20877606 | 0.91 |
ENSDART00000088106
|
ank3a
|
ankyrin 3a |
| chr13_+_44720106 | 0.91 |
ENSDART00000017770
|
zbtb8os
|
zinc finger and BTB domain containing 8 opposite strand |
| chr7_+_20215723 | 0.91 |
ENSDART00000173724
ENSDART00000173773 |
si:dkey-33c9.8
|
si:dkey-33c9.8 |
| chr3_+_47826386 | 0.91 |
ENSDART00000003338
|
unkl
|
unkempt family zinc finger-like |
| chr5_-_45905518 | 0.90 |
ENSDART00000111589
|
hapln1a
|
hyaluronan and proteoglycan link protein 1a |
| chr14_+_31564252 | 0.90 |
|
|
|
| chr23_+_2193582 | 0.89 |
ENSDART00000106336
|
cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr21_+_20734431 | 0.89 |
ENSDART00000079732
|
oxct1b
|
3-oxoacid CoA transferase 1b |
| chr25_-_13456748 | 0.89 |
ENSDART00000139290
|
ano10b
|
anoctamin 10b |
| chr17_-_29102320 | 0.88 |
ENSDART00000104204
|
foxg1a
|
forkhead box G1a |
| chr10_-_17630376 | 0.88 |
ENSDART00000113101
|
smarcad1b
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 b |
| chr13_+_22350043 | 0.88 |
ENSDART00000136863
|
ldb3a
|
LIM domain binding 3a |
| chr3_+_52448081 | 0.87 |
ENSDART00000154410
|
colgalt1
|
collagen beta(1-O)galactosyltransferase 1 |
| chr2_-_57133471 | 0.87 |
|
|
|
| chr5_+_58727940 | 0.87 |
ENSDART00000148659
|
clip2
|
CAP-GLY domain containing linker protein 2 |
| chr17_-_26893412 | 0.86 |
ENSDART00000045842
|
rcan3
|
regulator of calcineurin 3 |
| chr7_+_65532390 | 0.86 |
ENSDART00000156683
|
CT573494.2
|
ENSDARG00000097673 |
| chr19_-_44043682 | 0.86 |
ENSDART00000138009
ENSDART00000086138 |
fam83hb
|
family with sequence similarity 83, member Hb |
| chr23_+_36032206 | 0.85 |
ENSDART00000103132
|
hoxc4a
|
homeobox C4a |
| chr3_-_22082170 | 0.84 |
ENSDART00000155490
|
maptb
|
microtubule-associated protein tau b |
| chr18_+_48433986 | 0.84 |
ENSDART00000076861
|
fli1a
|
Fli-1 proto-oncogene, ETS transcription factor a |
| chr6_-_54816183 | 0.84 |
ENSDART00000148462
|
tnni1b
|
troponin I type 1b (skeletal, slow) |
| chr22_-_37468331 | 0.84 |
ENSDART00000160940
|
SOX2OT_exon1
|
SOX2 overlapping transcript exon 1 |
| chr7_-_27877036 | 0.83 |
ENSDART00000044208
|
lmo1
|
LIM domain only 1 |
| chr9_+_33167554 | 0.83 |
ENSDART00000007630
|
nhlh2
|
nescient helix loop helix 2 |
| chr3_-_49242655 | 0.83 |
ENSDART00000142200
ENSDART00000139524 |
arl16
|
ADP-ribosylation factor-like 16 |
| chr9_+_17341042 | 0.83 |
ENSDART00000147488
|
slain1a
|
SLAIN motif family, member 1a |
| chr2_-_32842855 | 0.82 |
ENSDART00000098834
|
prpf4ba
|
pre-mRNA processing factor 4Ba |
| chr19_-_43468872 | 0.82 |
|
|
|
| chr14_+_47326080 | 0.82 |
ENSDART00000047525
|
cryba1l1
|
crystallin, beta A1, like 1 |
| chr5_-_23495973 | 0.82 |
ENSDART00000085969
|
map7d2a
|
MAP7 domain containing 2a |
| chr23_+_29431173 | 0.81 |
ENSDART00000027255
|
tardbpl
|
TAR DNA binding protein, like |
| chr10_+_44195410 | 0.81 |
ENSDART00000046172
|
cryba4
|
crystallin, beta A4 |
| chr14_+_34683695 | 0.81 |
ENSDART00000170631
|
ebf1a
|
early B-cell factor 1a |
| chr3_+_14238188 | 0.80 |
ENSDART00000165452
ENSDART00000171726 |
tmem56b
|
transmembrane protein 56b |
| chr19_+_43523303 | 0.80 |
ENSDART00000167847
|
eef1a1l2
|
eukaryotic translation elongation factor 1 alpha 1, like 2 |
| chr4_-_824333 | 0.80 |
ENSDART00000135618
|
sobpb
|
sine oculis binding protein homolog (Drosophila) b |
| chr11_+_24720057 | 0.79 |
ENSDART00000145647
|
sulf2a
|
sulfatase 2a |
| chr12_-_3018686 | 0.79 |
ENSDART00000169161
|
dcxr
|
dicarbonyl/L-xylulose reductase |
| chr7_+_29969936 | 0.79 |
ENSDART00000109243
|
sema4bb
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Bb |
| chr19_+_341775 | 0.78 |
ENSDART00000151013
|
ensaa
|
endosulfine alpha a |
| chr3_+_24003840 | 0.78 |
ENSDART00000156204
|
si:ch211-246i5.5
|
si:ch211-246i5.5 |
| chr11_+_19894772 | 0.78 |
ENSDART00000103997
|
fezf2
|
FEZ family zinc finger 2 |
| chr8_+_16954190 | 0.78 |
ENSDART00000018934
|
pde4d
|
phosphodiesterase 4D, cAMP-specific |
| chr5_-_40310798 | 0.77 |
ENSDART00000133183
|
parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr14_-_4166292 | 0.77 |
ENSDART00000127318
|
frmpd1b
|
FERM and PDZ domain containing 1b |
| chr19_-_22184035 | 0.76 |
|
|
|
| chr15_-_17818432 | 0.76 |
ENSDART00000157198
|
CT573342.1
|
ENSDARG00000097333 |
| chr6_-_41087828 | 0.76 |
ENSDART00000028217
|
srsf3a
|
serine/arginine-rich splicing factor 3a |
| chr7_+_60054196 | 0.75 |
ENSDART00000039827
|
ppp1r14bb
|
protein phosphatase 1, regulatory (inhibitor) subunit 14Bb |
| chr6_-_43679553 | 0.75 |
ENSDART00000150128
|
foxp1b
|
forkhead box P1b |
| chr9_+_4188263 | 0.75 |
ENSDART00000164543
|
kalrna
|
kalirin RhoGEF kinase a |
| chr5_-_40310702 | 0.74 |
ENSDART00000133183
|
parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr24_-_40968409 | 0.74 |
ENSDART00000169315
ENSDART00000171543 |
smyhc1
|
slow myosin heavy chain 1 |
| chr3_-_38642067 | 0.74 |
ENSDART00000155518
|
znf281a
|
zinc finger protein 281a |
| chr6_-_39767452 | 0.73 |
ENSDART00000085277
|
pfkmb
|
phosphofructokinase, muscle b |
| chr16_+_5778650 | 0.73 |
ENSDART00000131575
|
zgc:158689
|
zgc:158689 |
| chr5_-_40310638 | 0.73 |
ENSDART00000133183
|
parp8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr17_-_25630666 | 0.73 |
ENSDART00000149060
|
ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isozyme |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:1990168 | protein K29-linked deubiquitination(GO:0035523) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) protein K33-linked deubiquitination(GO:1990168) |
| 0.6 | 1.9 | GO:0050810 | regulation of steroid biosynthetic process(GO:0050810) |
| 0.6 | 4.2 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.6 | 2.4 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.5 | 5.9 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) mitotic DNA replication(GO:1902969) |
| 0.5 | 5.0 | GO:0006991 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.5 | 1.8 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.4 | 1.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.4 | 2.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.4 | 2.5 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.3 | 2.0 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.3 | 2.6 | GO:0014831 | gastro-intestinal system smooth muscle contraction(GO:0014831) |
| 0.3 | 2.9 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.3 | 3.0 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) |
| 0.2 | 1.5 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.2 | 1.2 | GO:0048745 | smooth muscle tissue development(GO:0048745) arterial endothelial cell fate commitment(GO:0060844) |
| 0.2 | 0.9 | GO:1902224 | cellular ketone body metabolic process(GO:0046950) ketone body metabolic process(GO:1902224) |
| 0.2 | 0.7 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.2 | 2.0 | GO:0021767 | mammillary body development(GO:0021767) neuronal stem cell population maintenance(GO:0097150) |
| 0.2 | 0.9 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.2 | 1.2 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.2 | 1.2 | GO:0071322 | cellular response to carbohydrate stimulus(GO:0071322) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.2 | 1.1 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.2 | 1.7 | GO:0035108 | limb morphogenesis(GO:0035108) |
| 0.2 | 0.5 | GO:0032239 | regulation of nucleobase-containing compound transport(GO:0032239) positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of nucleocytoplasmic transport(GO:0046824) regulation of RNA export from nucleus(GO:0046831) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.2 | 0.7 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 0.2 | 1.0 | GO:0003232 | bulbus arteriosus development(GO:0003232) |
| 0.2 | 1.4 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.2 | 2.9 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.2 | 0.9 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.2 | 1.3 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 0.6 | GO:0003322 | pancreatic A cell development(GO:0003322) |
| 0.2 | 4.2 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.2 | 1.2 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 2.3 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.2 | 1.1 | GO:2000480 | regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.1 | 1.3 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.5 | GO:2000055 | positive regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000055) |
| 0.1 | 0.7 | GO:0042438 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.1 | 1.2 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.1 | 0.5 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.4 | GO:0061323 | cell proliferation involved in heart morphogenesis(GO:0061323) regulation of cell proliferation involved in heart morphogenesis(GO:2000136) |
| 0.1 | 1.6 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.1 | 1.0 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.1 | 2.7 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.1 | 1.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.1 | 0.6 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.1 | 0.8 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.1 | 0.7 | GO:0061718 | glucose catabolic process(GO:0006007) NADH regeneration(GO:0006735) glycolytic process through fructose-6-phosphate(GO:0061615) glycolytic process through glucose-6-phosphate(GO:0061620) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.8 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 1.3 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 0.6 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 1.2 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.1 | 0.3 | GO:0097090 | presynaptic membrane organization(GO:0097090) postsynaptic membrane assembly(GO:0097104) presynaptic membrane assembly(GO:0097105) |
| 0.1 | 0.9 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.1 | 0.7 | GO:0090109 | cell-substrate adherens junction assembly(GO:0007045) focal adhesion assembly(GO:0048041) regulation of focal adhesion assembly(GO:0051893) regulation of cell-substrate junction assembly(GO:0090109) |
| 0.1 | 1.0 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 0.9 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 5.1 | GO:0007601 | visual perception(GO:0007601) |
| 0.1 | 2.4 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.1 | 0.9 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.1 | 2.2 | GO:0021587 | cerebellum morphogenesis(GO:0021587) |
| 0.1 | 1.3 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 1.4 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 0.5 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.1 | 0.7 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.1 | 0.2 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 1.4 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.1 | 1.2 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.0 | 2.8 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.3 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 1.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.7 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.4 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 2.4 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 1.7 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 1.0 | GO:0030282 | bone mineralization(GO:0030282) |
| 0.0 | 0.3 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.8 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.7 | GO:0021854 | limbic system development(GO:0021761) hypothalamus development(GO:0021854) |
| 0.0 | 0.9 | GO:0006306 | DNA alkylation(GO:0006305) DNA methylation(GO:0006306) |
| 0.0 | 0.3 | GO:0050935 | iridophore differentiation(GO:0050935) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 1.1 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.7 | GO:0015696 | ammonium transport(GO:0015696) |
| 0.0 | 0.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.4 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0040019 | positive regulation of embryonic development(GO:0040019) |
| 0.0 | 1.3 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 2.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 1.4 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.1 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.0 | 0.6 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.7 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.5 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 0.9 | GO:0019722 | calcium-mediated signaling(GO:0019722) |
| 0.0 | 0.3 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.2 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.7 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.3 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.7 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.8 | GO:0006414 | translational elongation(GO:0006414) |
| 0.0 | 2.0 | GO:0001889 | liver development(GO:0001889) |
| 0.0 | 0.2 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.9 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) negative regulation of regulated secretory pathway(GO:1903306) |
| 0.0 | 3.0 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.0 | 1.3 | GO:0002244 | hematopoietic progenitor cell differentiation(GO:0002244) |
| 0.0 | 0.4 | GO:0031017 | exocrine pancreas development(GO:0031017) exocrine system development(GO:0035272) |
| 0.0 | 0.2 | GO:2001235 | positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.1 | GO:0016082 | synaptic vesicle priming(GO:0016082) |
| 0.0 | 0.4 | GO:0031638 | zymogen activation(GO:0031638) |
| 0.0 | 0.2 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.9 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.3 | 5.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 0.7 | GO:0071914 | prominosome(GO:0071914) |
| 0.2 | 1.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.2 | 0.7 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.2 | 3.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 0.9 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 2.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 0.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.0 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.7 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.5 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.1 | 1.0 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 0.5 | GO:0005915 | cell-cell adherens junction(GO:0005913) zonula adherens(GO:0005915) |
| 0.1 | 1.8 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.1 | 0.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 0.9 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 1.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 1.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 0.8 | GO:0031010 | ISWI-type complex(GO:0031010) |
| 0.1 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.3 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 0.3 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 2.4 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 7.9 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 3.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.9 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 1.5 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 2.2 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.9 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.5 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 2.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.4 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.2 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.3 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.0 | GO:0097433 | dense body(GO:0097433) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 5.9 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.6 | 2.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.6 | 2.4 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.6 | 2.9 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.5 | 2.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.5 | 2.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.4 | 3.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.4 | 1.8 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.3 | 1.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.3 | 1.3 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.3 | 1.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.2 | 1.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 0.9 | GO:0008410 | CoA-transferase activity(GO:0008410) |
| 0.2 | 0.9 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.2 | 6.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.2 | 3.2 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 2.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 1.1 | GO:0005536 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.2 | 0.6 | GO:0030504 | inorganic diphosphate transmembrane transporter activity(GO:0030504) |
| 0.2 | 1.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.2 | 0.9 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 0.7 | GO:0032034 | myosin head/neck binding(GO:0032028) myosin II head/neck binding(GO:0032034) myosin II heavy chain binding(GO:0032038) |
| 0.2 | 1.9 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.2 | 1.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 1.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 1.5 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.1 | 5.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 3.1 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.1 | 0.7 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.1 | 4.1 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 2.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.7 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 2.9 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 1.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.7 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) fructose-6-phosphate binding(GO:0070095) |
| 0.1 | 0.8 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.9 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 1.0 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.2 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.1 | 1.2 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.5 | GO:0005355 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.1 | 0.3 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.1 | 3.7 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 2.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.2 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.1 | 1.3 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.1 | 0.9 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.9 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.1 | 0.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.2 | GO:0019808 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 0.1 | 0.9 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 2.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.7 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.3 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 1.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.9 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 1.1 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.9 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0002039 | p53 binding(GO:0002039) |
| 0.0 | 2.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0051020 | GTPase binding(GO:0051020) |
| 0.0 | 0.2 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.0 | 0.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 1.9 | GO:0031406 | carboxylic acid binding(GO:0031406) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.9 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 22.2 | GO:0000987 | core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 1.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 2.3 | GO:0035091 | phosphatidylinositol binding(GO:0035091) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.4 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 0.8 | GO:0031625 | ubiquitin protein ligase binding(GO:0031625) |
| 0.0 | 3.2 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 1.4 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.5 | GO:0016860 | intramolecular oxidoreductase activity(GO:0016860) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 2.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 2.9 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.1 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 0.7 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.1 | 0.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.2 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.4 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 1.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.4 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 5.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.4 | 1.9 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.3 | 3.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.2 | 1.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.8 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 0.6 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 0.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.5 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.0 | 1.2 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.5 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.7 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.3 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.5 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 1.1 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |