Project
DANIO-CODE
Navigation
Downloads
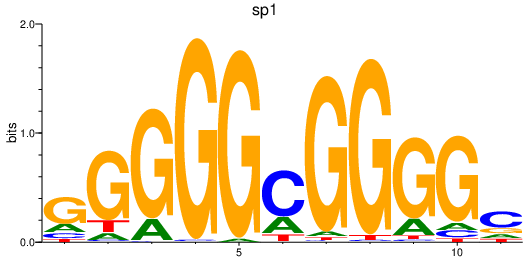
Results for sp1
Z-value: 4.19
Transcription factors associated with sp1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sp1
|
ENSDARG00000088347 | sp1 transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sp1 | dr10_dc_chr11_-_28193_28230 | -0.69 | 2.9e-03 | Click! |
Activity profile of sp1 motif
Sorted Z-values of sp1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of sp1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_+_33391554 | 14.47 |
ENSDART00000024104
|
mycn
|
MYCN proto-oncogene, bHLH transcription factor |
| chr13_+_11913290 | 12.65 |
ENSDART00000079398
|
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr16_-_46799406 | 11.61 |
ENSDART00000115244
|
MEX3A
|
mex-3 RNA binding family member A |
| chr5_-_43896815 | 10.95 |
ENSDART00000110076
|
gas1a
|
growth arrest-specific 1a |
| chr18_-_41660821 | 10.64 |
ENSDART00000024087
|
fzd9b
|
frizzled class receptor 9b |
| chr3_+_22412059 | 10.39 |
|
|
|
| chr19_-_32214978 | 10.04 |
ENSDART00000103640
|
hey1
|
hes-related family bHLH transcription factor with YRPW motif 1 |
| chr16_+_46327528 | 9.80 |
ENSDART00000040769
|
nr2f5
|
nuclear receptor subfamily 2, group F, member 5 |
| chr10_+_4094568 | 9.71 |
ENSDART00000114533
|
mn1a
|
meningioma 1a |
| chr22_+_13862110 | 9.46 |
ENSDART00000105711
|
sh3bp4a
|
SH3-domain binding protein 4a |
| chr3_+_16115708 | 9.43 |
ENSDART00000122519
|
st8sia6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
| chr16_-_7525980 | 9.27 |
ENSDART00000017445
|
prdm1a
|
PR domain containing 1a, with ZNF domain |
| chr6_-_39315024 | 9.13 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr2_+_11973282 | 9.03 |
ENSDART00000057268
|
greb1l
|
growth regulation by estrogen in breast cancer-like |
| chr5_+_4372660 | 8.54 |
ENSDART00000067599
|
angptl2a
|
angiopoietin-like 2a |
| chr3_+_32394653 | 8.43 |
ENSDART00000150897
|
si:ch73-367p23.2
|
si:ch73-367p23.2 |
| chr24_-_38068071 | 8.28 |
ENSDART00000041805
|
metrn
|
meteorin, glial cell differentiation regulator |
| chr7_+_34417030 | 8.23 |
ENSDART00000108473
|
plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr23_+_10412852 | 8.18 |
ENSDART00000142595
|
krt18
|
keratin 18 |
| chr12_-_26760324 | 8.14 |
ENSDART00000047724
|
zeb1b
|
zinc finger E-box binding homeobox 1b |
| chr18_-_11216447 | 8.14 |
ENSDART00000040500
|
tspan9a
|
tetraspanin 9a |
| chr22_+_10622013 | 8.08 |
ENSDART00000145459
|
rassf1
|
Ras association (RalGDS/AF-6) domain family 1 |
| chr5_-_43896757 | 8.01 |
ENSDART00000110076
|
gas1a
|
growth arrest-specific 1a |
| chr1_-_50066633 | 8.01 |
|
|
|
| chr11_+_25022419 | 7.99 |
|
|
|
| chr5_+_6054781 | 7.98 |
ENSDART00000060532
|
zgc:110796
|
zgc:110796 |
| chr15_+_19902697 | 7.87 |
ENSDART00000101204
|
alcamb
|
activated leukocyte cell adhesion molecule b |
| chr8_+_1122000 | 7.74 |
ENSDART00000127252
|
slc35d2
|
solute carrier family 35 (UDP-GlcNAc/UDP-glucose transporter), member D2 |
| chr22_-_35967525 | 7.72 |
ENSDART00000176971
|
CABZ01037120.1
|
ENSDARG00000106725 |
| chr11_-_471166 | 7.61 |
ENSDART00000154888
|
cnbpb
|
CCHC-type zinc finger, nucleic acid binding protein b |
| chr21_+_25199691 | 7.54 |
ENSDART00000168140
ENSDART00000112783 |
tmem45b
|
transmembrane protein 45B |
| chr7_+_21455138 | 7.51 |
ENSDART00000173992
|
kdm6ba
|
lysine (K)-specific demethylase 6B, a |
| chr18_-_49324015 | 7.48 |
ENSDART00000098419
|
sb:cb81
|
sb:cb81 |
| chr9_-_24431684 | 7.43 |
ENSDART00000039399
|
cavin2a
|
caveolae associated protein 2a |
| chr13_-_33696425 | 7.37 |
ENSDART00000143703
|
flrt3
|
fibronectin leucine rich transmembrane 3 |
| chr11_+_23522743 | 7.33 |
ENSDART00000121874
|
nfasca
|
neurofascin homolog (chicken) a |
| chr13_+_11912981 | 7.27 |
ENSDART00000158244
|
gng2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr1_-_18110990 | 7.25 |
ENSDART00000020970
|
pgm2
|
phosphoglucomutase 2 |
| chr11_-_32461160 | 7.21 |
ENSDART00000155592
|
pcdh17
|
protocadherin 17 |
| chr25_-_325847 | 7.15 |
ENSDART00000059514
|
prickle1a
|
prickle homolog 1a |
| chr20_-_25063937 | 7.08 |
ENSDART00000159122
|
epha7
|
eph receptor A7 |
| chr8_+_20125687 | 7.06 |
ENSDART00000124809
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr19_+_48582114 | 7.03 |
ENSDART00000157424
|
CU693379.1
|
ENSDARG00000098777 |
| chr3_-_53304694 | 6.96 |
ENSDART00000073930
|
notch3
|
notch 3 |
| chr7_-_23965038 | 6.93 |
ENSDART00000010124
|
slc7a8a
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8a |
| chr8_-_53508979 | 6.86 |
ENSDART00000158789
|
chdh
|
choline dehydrogenase |
| chr17_-_124685 | 6.82 |
ENSDART00000158825
|
actc1b
|
actin, alpha, cardiac muscle 1b |
| chr15_+_46114986 | 6.80 |
ENSDART00000156366
|
si:ch1073-340i21.3
|
si:ch1073-340i21.3 |
| chr10_-_33678206 | 6.80 |
ENSDART00000128049
|
hunk
|
hormonally up-regulated Neu-associated kinase |
| chr19_-_15951003 | 6.74 |
ENSDART00000133059
|
cited4a
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4a |
| chr3_-_62143464 | 6.73 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr22_+_30234718 | 6.70 |
ENSDART00000172496
|
add3a
|
adducin 3 (gamma) a |
| chr19_+_42491631 | 6.66 |
ENSDART00000150949
|
nfyc
|
nuclear transcription factor Y, gamma |
| chr5_-_7134405 | 6.65 |
ENSDART00000177935
|
bmpr1ba
|
bone morphogenetic protein receptor, type IBa |
| chr1_-_44888989 | 6.65 |
ENSDART00000135089
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr20_-_54192057 | 6.56 |
ENSDART00000023550
|
hsp90aa1.2
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 2 |
| chr16_-_24220413 | 6.55 |
ENSDART00000103176
|
bcam
|
basal cell adhesion molecule (Lutheran blood group) |
| chr23_-_24468191 | 6.54 |
ENSDART00000044918
|
epha2b
|
eph receptor A2 b |
| chr7_-_50827308 | 6.52 |
ENSDART00000121574
|
col4a6
|
collagen, type IV, alpha 6 |
| chr15_-_11257445 | 6.50 |
|
|
|
| chr20_+_30917073 | 6.48 |
|
|
|
| chr13_-_33574216 | 6.44 |
ENSDART00000065435
|
cst3
|
cystatin C (amyloid angiopathy and cerebral hemorrhage) |
| chr11_-_19612693 | 6.44 |
ENSDART00000037894
|
namptb
|
nicotinamide phosphoribosyltransferase b |
| chr23_-_45250886 | 6.44 |
ENSDART00000163367
|
ENSDARG00000103727
|
ENSDARG00000103727 |
| chr10_-_32580145 | 6.39 |
ENSDART00000137608
|
dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr22_+_11726312 | 6.35 |
ENSDART00000155366
|
krt96
|
keratin 96 |
| chr8_+_21163236 | 6.32 |
ENSDART00000091307
|
col2a1a
|
collagen, type II, alpha 1a |
| chr10_-_42032702 | 6.24 |
|
|
|
| chr12_+_689413 | 6.21 |
ENSDART00000174804
|
abcc3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
| chr3_+_28450576 | 6.17 |
ENSDART00000150893
|
sept12
|
septin 12 |
| chr23_+_36832325 | 6.13 |
|
|
|
| chr9_+_36051713 | 6.04 |
ENSDART00000134447
|
rcan1a
|
regulator of calcineurin 1a |
| chr4_-_25075693 | 6.01 |
ENSDART00000025153
|
gata3
|
GATA binding protein 3 |
| chr8_-_14142049 | 5.98 |
ENSDART00000126432
|
rhoaa
|
ras homolog gene family, member Aa |
| chr1_-_40536800 | 5.98 |
ENSDART00000134739
|
rgs12b
|
regulator of G protein signaling 12b |
| chr9_-_48098629 | 5.95 |
|
|
|
| chr14_+_4810147 | 5.94 |
ENSDART00000114678
|
nanos1
|
nanos homolog 1 |
| chr5_-_4091643 | 5.93 |
ENSDART00000067594
|
cst14b.1
|
cystatin 14b, tandem duplicate 1 |
| chr22_+_26543146 | 5.91 |
|
|
|
| chr24_-_1155215 | 5.91 |
ENSDART00000177356
|
itgb1a
|
integrin, beta 1a |
| chr8_+_584353 | 5.91 |
ENSDART00000048498
|
CABZ01049925.1
|
ENSDARG00000079790 |
| chr6_+_49096966 | 5.88 |
ENSDART00000141042
ENSDART00000143369 |
slc25a22
|
solute carrier family 25 (mitochondrial carrier: glutamate), member 22 |
| chr8_+_10267246 | 5.88 |
ENSDART00000159312
ENSDART00000160766 |
pim1
|
Pim-1 proto-oncogene, serine/threonine kinase |
| chr1_-_674449 | 5.86 |
ENSDART00000160564
|
cyyr1
|
cysteine/tyrosine-rich 1 |
| chr4_-_876235 | 5.86 |
|
|
|
| chr5_+_68410884 | 5.86 |
ENSDART00000153691
|
CU928129.1
|
ENSDARG00000097815 |
| chr13_-_22877318 | 5.86 |
ENSDART00000057638
ENSDART00000171778 |
hk1
|
hexokinase 1 |
| chr10_-_32580373 | 5.84 |
ENSDART00000066793
|
dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr19_-_11113032 | 5.82 |
ENSDART00000027598
|
tpm3
|
tropomyosin 3 |
| chr16_+_4825019 | 5.80 |
ENSDART00000167665
|
LIN28A
|
lin-28 homolog A |
| chr23_-_2573949 | 5.72 |
ENSDART00000123322
|
snai1b
|
snail family zinc finger 1b |
| chr3_+_5665300 | 5.72 |
ENSDART00000101807
|
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr21_+_5693372 | 5.70 |
ENSDART00000020603
|
ccng2
|
cyclin G2 |
| chr16_+_17705704 | 5.64 |
|
|
|
| chr21_+_25729090 | 5.62 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr5_-_7666964 | 5.59 |
ENSDART00000167643
|
slc1a3a
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3a |
| chr3_-_6078015 | 5.58 |
ENSDART00000165715
|
BX284638.1
|
ENSDARG00000098850 |
| chr21_-_32403403 | 5.58 |
ENSDART00000076974
|
gfpt2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr25_+_33527870 | 5.56 |
ENSDART00000011967
|
anxa2a
|
annexin A2a |
| chr10_+_15819127 | 5.53 |
ENSDART00000140511
|
apba1b
|
amyloid beta (A4) precursor protein-binding, family A, member 1b |
| chr5_-_18548689 | 5.53 |
ENSDART00000137802
|
fam214b
|
family with sequence similarity 214, member B |
| chr1_-_31123999 | 5.51 |
ENSDART00000152493
|
nt5c2b
|
5'-nucleotidase, cytosolic IIb |
| chr5_-_36647498 | 5.49 |
ENSDART00000112312
|
lrch2
|
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr1_+_29054033 | 5.48 |
ENSDART00000054066
|
zic2b
|
zic family member 2 (odd-paired homolog, Drosophila) b |
| chr6_-_2294751 | 5.42 |
ENSDART00000165223
|
pbx1b
|
pre-B-cell leukemia homeobox 1b |
| chr20_+_17840364 | 5.41 |
ENSDART00000024627
|
cdh2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr22_-_15567180 | 5.41 |
ENSDART00000123125
|
tpm4a
|
tropomyosin 4a |
| chr20_+_30894667 | 5.36 |
ENSDART00000145066
|
nhsl1b
|
NHS-like 1b |
| chr20_+_26408687 | 5.36 |
|
|
|
| chr11_-_44724371 | 5.34 |
ENSDART00000166501
|
cant1b
|
calcium activated nucleotidase 1b |
| chr17_+_45931793 | 5.34 |
ENSDART00000155372
|
kif26ab
|
kinesin family member 26Ab |
| chr15_-_33669717 | 5.34 |
ENSDART00000161151
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr4_+_1750689 | 5.31 |
ENSDART00000146779
|
slc38a2
|
solute carrier family 38, member 2 |
| chr9_-_9693163 | 5.27 |
ENSDART00000018228
|
gsk3b
|
glycogen synthase kinase 3 beta |
| chr21_-_39013447 | 5.26 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr16_-_32022177 | 5.25 |
ENSDART00000139664
|
styk1
|
serine/threonine/tyrosine kinase 1 |
| chr1_-_50513873 | 5.21 |
ENSDART00000137172
|
jag1a
|
jagged 1a |
| chr1_-_50831155 | 5.19 |
ENSDART00000152719
|
spred2a
|
sprouty-related, EVH1 domain containing 2a |
| chr18_-_45750 | 5.19 |
ENSDART00000148821
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr5_-_19357961 | 5.18 |
ENSDART00000088904
ENSDART00000163701 |
gltpa
|
glycolipid transfer protein a |
| chr5_-_40961381 | 5.17 |
|
|
|
| chr1_-_50513645 | 5.17 |
ENSDART00000047851
|
jag1a
|
jagged 1a |
| chr16_-_42486580 | 5.17 |
ENSDART00000148475
|
cspg5a
|
chondroitin sulfate proteoglycan 5a |
| chr10_-_28949111 | 5.17 |
ENSDART00000030138
|
alcama
|
activated leukocyte cell adhesion molecule a |
| chr19_-_24971633 | 5.16 |
ENSDART00000162801
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr19_+_12315359 | 5.14 |
ENSDART00000149221
|
grhl2b
|
grainyhead-like transcription factor 2b |
| chr15_-_21901138 | 5.13 |
ENSDART00000089953
|
sik2b
|
salt-inducible kinase 2b |
| chr23_+_44817648 | 5.12 |
ENSDART00000143688
|
dlg4b
|
discs, large homolog 4b (Drosophila) |
| chr1_-_18695214 | 5.08 |
|
|
|
| chr2_-_39710140 | 5.08 |
ENSDART00000147821
|
spsb4a
|
splA/ryanodine receptor domain and SOCS box containing 4a |
| chr23_-_7283514 | 5.08 |
ENSDART00000156369
|
CR589876.1
|
ENSDARG00000096997 |
| chr8_+_47861033 | 5.06 |
ENSDART00000147159
|
plod1a
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1a |
| chr19_-_7531709 | 5.03 |
ENSDART00000104750
|
mllt11
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 11 |
| chr13_+_22350043 | 5.03 |
ENSDART00000136863
|
ldb3a
|
LIM domain binding 3a |
| chr14_-_6901415 | 5.01 |
ENSDART00000167994
ENSDART00000166532 |
stox2b
|
storkhead box 2b |
| chr10_-_43815475 | 5.00 |
ENSDART00000027392
|
mef2ca
|
myocyte enhancer factor 2ca |
| chr18_+_38307946 | 5.00 |
ENSDART00000134247
|
lmo2
|
LIM domain only 2 (rhombotin-like 1) |
| chr15_-_4154407 | 4.98 |
ENSDART00000090624
|
lpar6a
|
lysophosphatidic acid receptor 6a |
| chr20_-_21773202 | 4.97 |
ENSDART00000133286
|
si:ch211-207i1.2
|
si:ch211-207i1.2 |
| chr21_-_36338514 | 4.97 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr17_+_15425559 | 4.97 |
ENSDART00000026180
|
fabp7a
|
fatty acid binding protein 7, brain, a |
| chr6_+_39373026 | 4.96 |
ENSDART00000157165
|
si:dkey-195m11.8
|
si:dkey-195m11.8 |
| chr2_-_24275851 | 4.96 |
ENSDART00000121885
|
tgfbr1a
|
transforming growth factor, beta receptor 1 a |
| chr2_-_43998886 | 4.92 |
ENSDART00000146493
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr16_+_42993841 | 4.88 |
ENSDART00000156767
|
CR854827.4
|
ENSDARG00000097750 |
| chr24_+_28364360 | 4.86 |
ENSDART00000018037
|
arhgap29a
|
Rho GTPase activating protein 29a |
| chr10_+_10830549 | 4.85 |
ENSDART00000101077
ENSDART00000139143 |
ptgdsa
|
prostaglandin D2 synthase a |
| chr11_-_42934175 | 4.85 |
ENSDART00000172929
|
sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr20_-_158899 | 4.85 |
ENSDART00000131635
|
slc16a10
|
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr7_+_73408688 | 4.85 |
ENSDART00000159745
|
PCP4L1
|
Purkinje cell protein 4 like 1 |
| chr11_-_25693072 | 4.81 |
ENSDART00000122011
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr12_+_11042472 | 4.79 |
ENSDART00000079336
|
raraa
|
retinoic acid receptor, alpha a |
| chr19_+_31046291 | 4.79 |
ENSDART00000052124
|
fam49al
|
family with sequence similarity 49, member A-like |
| chr24_-_21114505 | 4.69 |
ENSDART00000111025
|
boc
|
BOC cell adhesion associated, oncogene regulated |
| chr21_-_11554253 | 4.67 |
ENSDART00000133443
|
cast
|
calpastatin |
| chr19_-_1905372 | 4.63 |
ENSDART00000029646
|
rplp1
|
ribosomal protein, large, P1 |
| chr15_-_9026081 | 4.59 |
ENSDART00000126708
|
rhoub
|
ras homolog family member Ub |
| chr5_-_56984800 | 4.59 |
ENSDART00000074290
|
mia
|
melanoma inhibitory activity |
| chr13_+_22173410 | 4.58 |
ENSDART00000173405
|
usp54a
|
ubiquitin specific peptidase 54a |
| chr20_-_23184142 | 4.57 |
ENSDART00000176282
|
CU693368.2
|
ENSDARG00000108718 |
| chr7_-_64637419 | 4.56 |
ENSDART00000020456
|
mmp15b
|
matrix metallopeptidase 15b |
| chr7_-_24093289 | 4.56 |
ENSDART00000064789
|
txn
|
thioredoxin |
| chr19_-_5338129 | 4.54 |
ENSDART00000081951
|
stx1b
|
syntaxin 1B |
| chr8_+_4641405 | 4.54 |
ENSDART00000064197
|
cldn5a
|
claudin 5a |
| chr23_+_45020761 | 4.53 |
ENSDART00000159104
|
atp1b2a
|
ATPase, Na+/K+ transporting, beta 2a polypeptide |
| chr18_+_26102262 | 4.52 |
ENSDART00000164495
|
znf710a
|
zinc finger protein 710a |
| chr3_+_15656123 | 4.52 |
ENSDART00000055834
|
phospho1
|
phosphatase, orphan 1 |
| chr14_-_40454194 | 4.51 |
ENSDART00000166621
|
ef1
|
E74-like factor 1 (ets domain transcription factor) |
| chr7_-_16346150 | 4.48 |
ENSDART00000128488
|
e2f8
|
E2F transcription factor 8 |
| chr9_-_22140954 | 4.48 |
ENSDART00000146528
|
lmo7a
|
LIM domain 7a |
| chr3_+_16115532 | 4.47 |
|
|
|
| chr19_-_617298 | 4.47 |
ENSDART00000171387
|
cyp51
|
cytochrome P450, family 51 |
| chr5_-_39910235 | 4.46 |
ENSDART00000146237
ENSDART00000163302 |
fsta
|
follistatin a |
| chr3_+_52901602 | 4.45 |
ENSDART00000114343
|
brd4
|
bromodomain containing 4 |
| chr2_-_38242982 | 4.43 |
ENSDART00000040357
|
acin1a
|
apoptotic chromatin condensation inducer 1a |
| chr1_-_416138 | 4.42 |
ENSDART00000092524
|
rasa3
|
RAS p21 protein activator 3 |
| chr13_+_4276783 | 4.39 |
ENSDART00000148280
|
prr18
|
proline rich 18 |
| chr24_-_4418454 | 4.38 |
ENSDART00000066835
|
fzd8a
|
frizzled class receptor 8a |
| chr20_-_53560663 | 4.36 |
ENSDART00000146001
|
wasf1
|
WAS protein family, member 1 |
| chr10_+_404464 | 4.36 |
ENSDART00000145124
ENSDART00000124319 |
dact3a
|
dishevelled-binding antagonist of beta-catenin 3a |
| KN150495v1_-_20497 | 4.35 |
|
|
|
| chr21_-_43806271 | 4.34 |
ENSDART00000138710
|
pdlim4
|
PDZ and LIM domain 4 |
| chr14_-_2682064 | 4.34 |
ENSDART00000161677
|
si:dkey-201i24.6
|
si:dkey-201i24.6 |
| chr23_-_28420470 | 4.33 |
ENSDART00000145072
|
neurod4
|
neuronal differentiation 4 |
| chr15_-_20532537 | 4.32 |
ENSDART00000018514
|
dlc
|
deltaC |
| chr8_+_6989445 | 4.31 |
ENSDART00000134440
|
gpd1a
|
glycerol-3-phosphate dehydrogenase 1a |
| chr14_+_21531709 | 4.30 |
ENSDART00000144367
|
ctbp1
|
C-terminal binding protein 1 |
| chr5_-_65349550 | 4.26 |
ENSDART00000164228
|
nrarpb
|
notch-regulated ankyrin repeat protein b |
| chr11_-_28367271 | 4.24 |
ENSDART00000065853
|
dhrs3b
|
dehydrogenase/reductase (SDR family) member 3b |
| chr10_+_9716807 | 4.24 |
ENSDART00000064977
|
si:ch211-243g18.2
|
si:ch211-243g18.2 |
| chr18_+_26437604 | 4.24 |
ENSDART00000060245
|
ctsh
|
cathepsin H |
| chr8_-_24991984 | 4.23 |
ENSDART00000078795
|
ahcyl1
|
adenosylhomocysteinase-like 1 |
| chr1_+_54231447 | 4.23 |
ENSDART00000145652
|
golga7ba
|
golgin A7 family, member Ba |
| chr5_+_53854279 | 4.22 |
ENSDART00000165889
|
tprn
|
taperin |
| chr3_+_12401750 | 4.20 |
ENSDART00000167177
|
ccnf
|
cyclin F |
| chr7_+_44373815 | 4.19 |
ENSDART00000170721
|
si:dkey-56m19.5
|
si:dkey-56m19.5 |
| chr6_+_2849460 | 4.18 |
|
|
|
| chr20_+_37940880 | 4.16 |
ENSDART00000165887
|
flvcr1
|
feline leukemia virus subgroup C cellular receptor 1 |
| chr4_+_3344998 | 4.15 |
ENSDART00000075320
|
nampta
|
nicotinamide phosphoribosyltransferase a |
| chr23_+_24141576 | 4.15 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.4 | 13.1 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 4.1 | 12.2 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 2.9 | 8.8 | GO:0006600 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 2.3 | 6.8 | GO:0072025 | distal convoluted tubule development(GO:0072025) late distal convoluted tubule development(GO:0072068) |
| 2.1 | 4.3 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 2.0 | 6.1 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) positive regulation of DNA endoreduplication(GO:0032877) DNA endoreduplication(GO:0042023) |
| 1.8 | 12.3 | GO:0021628 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 1.7 | 7.0 | GO:0035633 | maintenance of blood-brain barrier(GO:0035633) |
| 1.7 | 8.6 | GO:0046677 | response to antibiotic(GO:0046677) |
| 1.7 | 5.2 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 1.7 | 5.1 | GO:0072578 | neurotransmitter-gated ion channel clustering(GO:0072578) |
| 1.7 | 5.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 1.7 | 6.7 | GO:0099565 | chemical synaptic transmission, postsynaptic(GO:0099565) |
| 1.7 | 9.9 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) |
| 1.6 | 8.1 | GO:0045582 | positive regulation of T cell differentiation(GO:0045582) |
| 1.6 | 4.7 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 1.5 | 6.1 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 1.5 | 4.5 | GO:0098881 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 1.5 | 10.4 | GO:0060088 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 1.5 | 7.4 | GO:0051899 | membrane depolarization(GO:0051899) |
| 1.5 | 4.4 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 1.4 | 8.5 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 1.4 | 4.2 | GO:0097037 | heme export(GO:0097037) |
| 1.4 | 4.1 | GO:0009957 | epidermal cell fate specification(GO:0009957) cranial ganglion development(GO:0061550) |
| 1.3 | 5.4 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 1.3 | 10.8 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 1.3 | 3.9 | GO:0033690 | positive regulation of osteoblast proliferation(GO:0033690) |
| 1.3 | 3.9 | GO:1904398 | regulation of synaptic growth at neuromuscular junction(GO:0008582) axon regeneration at neuromuscular junction(GO:0014814) positive regulation of synaptic growth at neuromuscular junction(GO:0045887) positive regulation of neuromuscular junction development(GO:1904398) |
| 1.3 | 5.2 | GO:0010801 | regulation of peptidyl-threonine phosphorylation(GO:0010799) negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 1.3 | 7.6 | GO:2000767 | positive regulation of cytoplasmic translation(GO:2000767) |
| 1.3 | 6.3 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 1.2 | 3.7 | GO:0014821 | phasic smooth muscle contraction(GO:0014821) |
| 1.2 | 3.7 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 1.2 | 11.0 | GO:0001709 | cell fate determination(GO:0001709) |
| 1.2 | 2.4 | GO:0018197 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 1.2 | 8.3 | GO:0019226 | transmission of nerve impulse(GO:0019226) |
| 1.1 | 3.4 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 1.1 | 7.9 | GO:0021885 | forebrain cell migration(GO:0021885) |
| 1.1 | 28.8 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 1.1 | 3.3 | GO:0097435 | fibril organization(GO:0097435) |
| 1.1 | 4.3 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 1.1 | 3.2 | GO:0061614 | pri-miRNA transcription from RNA polymerase II promoter(GO:0061614) regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902893) positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 1.1 | 7.4 | GO:0006868 | glutamine transport(GO:0006868) |
| 1.1 | 3.2 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 1.0 | 5.2 | GO:0048795 | swim bladder morphogenesis(GO:0048795) |
| 1.0 | 17.1 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 1.0 | 4.0 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 1.0 | 6.9 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 1.0 | 2.9 | GO:0002706 | regulation of lymphocyte mediated immunity(GO:0002706) |
| 1.0 | 7.6 | GO:0045667 | regulation of osteoblast differentiation(GO:0045667) |
| 0.9 | 7.1 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.9 | 4.3 | GO:0001839 | neural plate morphogenesis(GO:0001839) |
| 0.9 | 4.3 | GO:0006116 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.9 | 9.5 | GO:0032986 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.9 | 2.6 | GO:0032656 | interleukin-13 production(GO:0032616) regulation of interleukin-13 production(GO:0032656) |
| 0.8 | 4.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.8 | 2.5 | GO:0070983 | dendrite guidance(GO:0070983) regulation of homophilic cell adhesion(GO:1903385) |
| 0.8 | 4.8 | GO:0007343 | egg activation(GO:0007343) |
| 0.8 | 9.4 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.8 | 3.9 | GO:0002455 | humoral immune response mediated by circulating immunoglobulin(GO:0002455) complement activation, classical pathway(GO:0006958) |
| 0.8 | 9.2 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.7 | 3.7 | GO:0048635 | negative regulation of muscle organ development(GO:0048635) |
| 0.7 | 15.1 | GO:0030878 | thyroid gland development(GO:0030878) |
| 0.7 | 5.0 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.7 | 2.8 | GO:0000050 | urea cycle(GO:0000050) urea metabolic process(GO:0019627) |
| 0.7 | 5.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.7 | 2.1 | GO:0072045 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) convergent extension involved in nephron morphogenesis(GO:0072045) |
| 0.7 | 2.1 | GO:0010586 | miRNA metabolic process(GO:0010586) |
| 0.7 | 5.5 | GO:0033278 | cell proliferation in midbrain(GO:0033278) |
| 0.7 | 2.7 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.7 | 2.0 | GO:0006478 | protein sulfation(GO:0006477) peptidyl-tyrosine sulfation(GO:0006478) |
| 0.7 | 2.0 | GO:0050748 | N-terminal protein palmitoylation(GO:0006500) negative regulation of lipoprotein metabolic process(GO:0050748) regulation of N-terminal protein palmitoylation(GO:0060254) negative regulation of N-terminal protein palmitoylation(GO:0060262) negative regulation of protein lipidation(GO:1903060) |
| 0.7 | 2.0 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.7 | 6.5 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.6 | 3.9 | GO:0043620 | regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) regulation of DNA-templated transcription in response to stress(GO:0043620) |
| 0.6 | 7.1 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.6 | 3.2 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.6 | 22.9 | GO:0050772 | positive regulation of axonogenesis(GO:0050772) |
| 0.6 | 8.1 | GO:0030574 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.6 | 1.8 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.6 | 4.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.6 | 1.8 | GO:0019835 | cytolysis(GO:0019835) |
| 0.6 | 3.5 | GO:0045670 | regulation of osteoclast differentiation(GO:0045670) |
| 0.6 | 2.3 | GO:0006751 | glutathione catabolic process(GO:0006751) response to estradiol(GO:0032355) |
| 0.6 | 2.9 | GO:0061162 | establishment of apical/basal cell polarity(GO:0035089) establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.6 | 3.5 | GO:0010960 | magnesium ion homeostasis(GO:0010960) |
| 0.6 | 1.1 | GO:1901379 | regulation of potassium ion transport(GO:0043266) regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.5 | 21.9 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.5 | 6.0 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.5 | 7.4 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) blood vessel lumenization(GO:0072554) |
| 0.5 | 4.7 | GO:0097341 | inhibition of cysteine-type endopeptidase activity(GO:0097340) zymogen inhibition(GO:0097341) |
| 0.5 | 4.6 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.5 | 3.1 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.5 | 9.1 | GO:0071711 | basement membrane organization(GO:0071711) |
| 0.5 | 4.5 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.5 | 6.0 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.5 | 6.9 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.5 | 7.4 | GO:1903672 | positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.5 | 3.9 | GO:0033555 | multicellular organismal response to stress(GO:0033555) |
| 0.5 | 2.8 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.5 | 2.3 | GO:0072512 | ferric iron transport(GO:0015682) transferrin transport(GO:0033572) trivalent inorganic cation transport(GO:0072512) |
| 0.5 | 0.5 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.5 | 6.8 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.5 | 1.4 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.5 | 5.9 | GO:0060561 | apoptotic process involved in morphogenesis(GO:0060561) |
| 0.4 | 2.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.4 | 10.2 | GO:0007632 | visual behavior(GO:0007632) |
| 0.4 | 1.7 | GO:0021703 | locus ceruleus development(GO:0021703) |
| 0.4 | 2.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.4 | 5.2 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.4 | 6.4 | GO:0051149 | positive regulation of myoblast differentiation(GO:0045663) positive regulation of muscle cell differentiation(GO:0051149) |
| 0.4 | 7.1 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.4 | 2.9 | GO:0006584 | catecholamine metabolic process(GO:0006584) catechol-containing compound metabolic process(GO:0009712) |
| 0.4 | 0.8 | GO:0002762 | negative regulation of myeloid leukocyte differentiation(GO:0002762) regulation of granulocyte differentiation(GO:0030852) regulation of neutrophil differentiation(GO:0045658) |
| 0.4 | 4.3 | GO:0010770 | positive regulation of cell morphogenesis involved in differentiation(GO:0010770) |
| 0.4 | 7.4 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.4 | 9.3 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.4 | 1.5 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.4 | 2.3 | GO:0010332 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.4 | 4.6 | GO:0044247 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.4 | 3.0 | GO:0048796 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.4 | 4.9 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.4 | 4.5 | GO:0001678 | cellular glucose homeostasis(GO:0001678) |
| 0.4 | 6.3 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.4 | 2.6 | GO:0070650 | actin filament bundle distribution(GO:0070650) |
| 0.4 | 0.7 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.3 | 6.6 | GO:0050654 | chondroitin sulfate proteoglycan metabolic process(GO:0050654) |
| 0.3 | 7.2 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.3 | 1.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.3 | 2.0 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.3 | 6.7 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.3 | 2.3 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.3 | 1.3 | GO:0035881 | amacrine cell differentiation(GO:0035881) |
| 0.3 | 2.0 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.3 | 8.7 | GO:0060119 | inner ear receptor cell development(GO:0060119) |
| 0.3 | 2.0 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.3 | 3.6 | GO:0060021 | palate development(GO:0060021) |
| 0.3 | 1.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 4.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.3 | 3.9 | GO:0061099 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.3 | 3.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.3 | 1.6 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.3 | 5.9 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.3 | 2.8 | GO:1902254 | regulation of retinal ganglion cell axon guidance(GO:0090259) negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.3 | 4.6 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 16.2 | GO:0035567 | non-canonical Wnt signaling pathway(GO:0035567) |
| 0.3 | 1.5 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.3 | 3.5 | GO:0046620 | regulation of organ growth(GO:0046620) |
| 0.3 | 3.5 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.3 | 4.6 | GO:0060319 | primitive erythrocyte differentiation(GO:0060319) |
| 0.3 | 6.0 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.3 | 3.8 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.3 | 1.3 | GO:0051177 | meiotic sister chromatid segregation(GO:0045144) meiotic sister chromatid cohesion(GO:0051177) meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.3 | 3.2 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.3 | 0.8 | GO:0070278 | extracellular matrix constituent secretion(GO:0070278) |
| 0.3 | 2.7 | GO:0061337 | cardiac conduction(GO:0061337) |
| 0.3 | 1.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.3 | 2.9 | GO:0090311 | regulation of histone deacetylation(GO:0031063) regulation of protein deacetylation(GO:0090311) |
| 0.2 | 1.7 | GO:0072160 | pronephric nephron tubule epithelial cell differentiation(GO:0035778) cell differentiation involved in pronephros development(GO:0039014) nephron tubule epithelial cell differentiation(GO:0072160) |
| 0.2 | 4.2 | GO:0048382 | mesendoderm development(GO:0048382) |
| 0.2 | 2.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.2 | 1.7 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.2 | 20.9 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.2 | 1.5 | GO:0061075 | positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) |
| 0.2 | 4.8 | GO:0030032 | lamellipodium assembly(GO:0030032) lamellipodium organization(GO:0097581) |
| 0.2 | 9.4 | GO:0030835 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) actin filament capping(GO:0051693) |
| 0.2 | 0.2 | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) |
| 0.2 | 4.9 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.2 | 1.4 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.2 | 0.2 | GO:0042059 | negative regulation of epidermal growth factor receptor signaling pathway(GO:0042059) negative regulation of ERBB signaling pathway(GO:1901185) |
| 0.2 | 2.5 | GO:0046686 | response to cadmium ion(GO:0046686) |
| 0.2 | 5.5 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.2 | 3.5 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.2 | 5.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.2 | 1.9 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.2 | 1.3 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.2 | 0.4 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.2 | 1.5 | GO:1902914 | negative regulation of histone ubiquitination(GO:0033183) regulation of protein K63-linked ubiquitination(GO:1900044) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) regulation of protein polyubiquitination(GO:1902914) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.2 | 0.9 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.2 | 5.6 | GO:0045666 | positive regulation of neuron differentiation(GO:0045666) |
| 0.2 | 5.5 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.2 | 4.5 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.2 | 2.0 | GO:0030168 | platelet activation(GO:0030168) |
| 0.2 | 0.6 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.2 | 3.4 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.2 | 4.2 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.2 | 2.0 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.2 | 0.6 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.2 | 1.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 0.8 | GO:0048339 | paraxial mesoderm development(GO:0048339) |
| 0.2 | 9.9 | GO:0006414 | translational elongation(GO:0006414) |
| 0.2 | 0.6 | GO:0060832 | oocyte animal/vegetal axis specification(GO:0060832) |
| 0.2 | 0.6 | GO:0035776 | pronephric proximal tubule development(GO:0035776) |
| 0.2 | 1.9 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.2 | 1.5 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.2 | 3.5 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.2 | 2.4 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.2 | 1.8 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.2 | 3.6 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.2 | 3.6 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 3.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.2 | 3.5 | GO:0014866 | skeletal myofibril assembly(GO:0014866) |
| 0.2 | 0.7 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.2 | 1.4 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.2 | 1.0 | GO:2001106 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 2.3 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.2 | 10.2 | GO:0001708 | cell fate specification(GO:0001708) |
| 0.2 | 1.0 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.2 | 2.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.2 | 1.8 | GO:0097178 | ruffle organization(GO:0031529) ruffle assembly(GO:0097178) |
| 0.2 | 0.5 | GO:0046655 | folic acid metabolic process(GO:0046655) |
| 0.1 | 0.6 | GO:0022900 | electron transport chain(GO:0022900) |
| 0.1 | 1.5 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 0.6 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 0.9 | GO:0008210 | estrogen biosynthetic process(GO:0006703) estrogen metabolic process(GO:0008210) |
| 0.1 | 5.4 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.1 | 0.4 | GO:0098773 | skin epidermis development(GO:0098773) |
| 0.1 | 13.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.1 | 0.1 | GO:1904357 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.1 | 1.6 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.7 | GO:0010990 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 4.2 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.1 | 0.4 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) |
| 0.1 | 1.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.9 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.1 | 0.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 1.0 | GO:0035305 | negative regulation of dephosphorylation(GO:0035305) negative regulation of protein dephosphorylation(GO:0035308) |
| 0.1 | 3.8 | GO:1903844 | regulation of transforming growth factor beta receptor signaling pathway(GO:0017015) regulation of cellular response to transforming growth factor beta stimulus(GO:1903844) |
| 0.1 | 0.5 | GO:0008592 | regulation of Toll signaling pathway(GO:0008592) |
| 0.1 | 1.4 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.9 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) |
| 0.1 | 1.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.7 | GO:0072576 | liver morphogenesis(GO:0072576) |
| 0.1 | 0.7 | GO:0007622 | rhythmic behavior(GO:0007622) circadian behavior(GO:0048512) |
| 0.1 | 6.2 | GO:0030155 | regulation of cell adhesion(GO:0030155) |
| 0.1 | 0.4 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) protein insertion into mitochondrial membrane from inner side(GO:0032979) protein insertion into mitochondrial membrane(GO:0051204) |
| 0.1 | 2.0 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.1 | 0.6 | GO:1902624 | positive regulation of neutrophil migration(GO:1902624) |
| 0.1 | 1.9 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.1 | 4.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.1 | 1.4 | GO:0036465 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) |
| 0.1 | 0.8 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.1 | 6.2 | GO:0048565 | digestive tract development(GO:0048565) |
| 0.1 | 0.9 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 1.1 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 1.3 | GO:0006094 | gluconeogenesis(GO:0006094) |
| 0.1 | 6.4 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.1 | 0.9 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 1.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 3.5 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.1 | 2.0 | GO:0060326 | cell chemotaxis(GO:0060326) |
| 0.1 | 0.4 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.1 | 2.4 | GO:0070509 | calcium ion import(GO:0070509) |
| 0.1 | 1.5 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 1.3 | GO:0043627 | response to estrogen(GO:0043627) |
| 0.1 | 1.7 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.1 | 3.6 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.1 | 1.0 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.1 | 2.6 | GO:1902600 | hydrogen ion transmembrane transport(GO:1902600) |
| 0.1 | 0.7 | GO:0019471 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.1 | 0.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 2.5 | GO:0031032 | actomyosin structure organization(GO:0031032) |
| 0.1 | 0.2 | GO:0070498 | interleukin-1-mediated signaling pathway(GO:0070498) |
| 0.1 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 2.1 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.1 | 1.1 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.1 | 6.0 | GO:0007411 | axon guidance(GO:0007411) |
| 0.1 | 0.8 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 2.9 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.1 | 2.2 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.1 | 6.1 | GO:0060548 | negative regulation of cell death(GO:0060548) |
| 0.1 | 0.6 | GO:0006911 | phagocytosis, engulfment(GO:0006911) |
| 0.1 | 5.7 | GO:0007268 | chemical synaptic transmission(GO:0007268) anterograde trans-synaptic signaling(GO:0098916) |
| 0.1 | 1.1 | GO:0007041 | lysosomal transport(GO:0007041) |
| 0.1 | 0.9 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.5 | GO:0032438 | melanosome organization(GO:0032438) |
| 0.0 | 0.3 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.0 | 0.8 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.7 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 4.2 | GO:0006352 | DNA-templated transcription, initiation(GO:0006352) |
| 0.0 | 3.6 | GO:0045333 | cellular respiration(GO:0045333) |
| 0.0 | 0.8 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 2.9 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 0.3 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.0 | 0.5 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.8 | GO:0006909 | phagocytosis(GO:0006909) |
| 0.0 | 0.9 | GO:0030705 | cytoskeleton-dependent intracellular transport(GO:0030705) |
| 0.0 | 0.6 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.0 | 0.5 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.0 | 0.3 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.8 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.6 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 1.2 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.4 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.2 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.0 | 3.3 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.0 | 1.1 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.0 | 1.9 | GO:0048024 | regulation of mRNA splicing, via spliceosome(GO:0048024) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.3 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:0098969 | neurotransmitter receptor transport to plasma membrane(GO:0098877) neurotransmitter receptor transport to postsynaptic membrane(GO:0098969) establishment of protein localization to postsynaptic membrane(GO:1903540) |
| 0.0 | 0.4 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 20.7 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 1.8 | 9.2 | GO:0008091 | spectrin(GO:0008091) |
| 1.7 | 6.7 | GO:0070062 | extracellular exosome(GO:0070062) |
| 1.6 | 6.5 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 1.2 | 6.0 | GO:0098890 | extrinsic component of postsynaptic membrane(GO:0098890) |
| 1.2 | 14.3 | GO:0043209 | myelin sheath(GO:0043209) |
| 1.2 | 9.2 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 1.0 | 10.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.9 | 8.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.9 | 2.6 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.8 | 5.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.8 | 3.9 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.7 | 4.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.7 | 3.5 | GO:0048788 | cytoskeleton of presynaptic active zone(GO:0048788) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.7 | 11.1 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.7 | 4.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.7 | 7.9 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.7 | 2.0 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.6 | 7.7 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.6 | 7.6 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.5 | 6.8 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.5 | 7.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.5 | 14.3 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.5 | 38.1 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.4 | 4.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.4 | 5.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.4 | 4.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.4 | 19.7 | GO:0042641 | actomyosin(GO:0042641) |
| 0.3 | 17.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.3 | 2.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.3 | 2.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.3 | 7.4 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.3 | 1.6 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.3 | 4.6 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.3 | 7.0 | GO:0032156 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.3 | 1.8 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.3 | 5.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 15.6 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.3 | 10.7 | GO:0005604 | basement membrane(GO:0005604) extracellular matrix component(GO:0044420) |
| 0.3 | 1.0 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.3 | 8.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.2 | 9.9 | GO:0005905 | clathrin-coated pit(GO:0005905) |
| 0.2 | 8.2 | GO:0005901 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.2 | 1.1 | GO:0032420 | stereocilium(GO:0032420) |
| 0.2 | 1.5 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.2 | 8.4 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.2 | 3.3 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.2 | 1.2 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.2 | 2.9 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 9.8 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.2 | 1.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.2 | 6.5 | GO:0099572 | postsynaptic specialization(GO:0099572) |
| 0.2 | 1.2 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.2 | 32.5 | GO:0043235 | receptor complex(GO:0043235) |
| 0.2 | 5.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.2 | 21.1 | GO:0030424 | axon(GO:0030424) |
| 0.2 | 3.9 | GO:0060293 | germ plasm(GO:0060293) |
| 0.2 | 1.7 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 6.7 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 4.6 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.8 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 4.9 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.1 | 1.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 1.1 | GO:0034705 | voltage-gated potassium channel complex(GO:0008076) potassium channel complex(GO:0034705) |
| 0.1 | 1.6 | GO:0000145 | exocyst(GO:0000145) |
| 0.1 | 2.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.1 | 51.6 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 2.3 | GO:0030426 | growth cone(GO:0030426) |
| 0.1 | 3.0 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.1 | 3.9 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 0.2 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.1 | 1.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 3.1 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.1 | 49.2 | GO:0005615 | extracellular space(GO:0005615) |
| 0.1 | 4.6 | GO:0005776 | autophagosome(GO:0005776) |
| 0.1 | 1.6 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 9.4 | GO:0005912 | adherens junction(GO:0005912) anchoring junction(GO:0070161) |
| 0.1 | 1.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.5 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 3.3 | GO:0022626 | cytosolic ribosome(GO:0022626) cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 5.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 1.4 | GO:0043025 | neuronal cell body(GO:0043025) |
| 0.1 | 4.7 | GO:0070382 | exocytic vesicle(GO:0070382) |
| 0.1 | 2.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.1 | 2.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.1 | 0.9 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.1 | 2.7 | GO:0005884 | actin filament(GO:0005884) |
| 0.1 | 3.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.1 | 5.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 2.2 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.0 | 2.5 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 3.7 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.8 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 1.3 | GO:0005865 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.7 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.6 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 86.0 | GO:0016021 | integral component of membrane(GO:0016021) |
| 0.0 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 4.0 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.8 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.3 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 2.4 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.6 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.7 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.5 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 8.0 | GO:0070548 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity(GO:0004360) L-glutamine aminotransferase activity(GO:0070548) |
| 2.6 | 7.8 | GO:0098639 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 2.6 | 7.7 | GO:0005463 | UDP-glucuronic acid transmembrane transporter activity(GO:0005461) UDP-N-acetylgalactosamine transmembrane transporter activity(GO:0005463) |
| 1.7 | 20.7 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 1.6 | 4.8 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 1.5 | 12.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 1.4 | 24.7 | GO:0005112 | Notch binding(GO:0005112) |
| 1.3 | 10.8 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 1.3 | 6.7 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 1.3 | 7.9 | GO:0016801 | hydrolase activity, acting on ether bonds(GO:0016801) |
| 1.3 | 18.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 1.2 | 5.0 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 1.1 | 12.3 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 1.1 | 2.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 1.1 | 6.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 1.1 | 20.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 1.1 | 4.3 | GO:0030274 | LIM domain binding(GO:0030274) |
| 1.1 | 3.2 | GO:0004953 | icosanoid receptor activity(GO:0004953) |
| 1.0 | 4.1 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 1.0 | 3.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 1.0 | 5.0 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 1.0 | 2.9 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 1.0 | 5.9 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) glucose binding(GO:0005536) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 1.0 | 2.9 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 1.0 | 4.8 | GO:0072349 | modified amino acid transmembrane transporter activity(GO:0072349) |
| 1.0 | 6.7 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.9 | 2.8 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.9 | 12.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.9 | 3.7 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.9 | 4.6 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.9 | 7.9 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.9 | 4.3 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.9 | 0.9 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.8 | 7.4 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.8 | 4.9 | GO:0019956 | chemokine binding(GO:0019956) C-C chemokine binding(GO:0019957) |
| 0.8 | 2.4 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.8 | 10.9 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.8 | 2.3 | GO:0000048 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.8 | 3.9 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.8 | 3.1 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.7 | 6.5 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.7 | 5.6 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.7 | 5.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.7 | 2.7 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.7 | 4.0 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.7 | 6.7 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.7 | 2.0 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.7 | 6.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.7 | 2.0 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.7 | 2.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.7 | 5.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.6 | 2.6 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.6 | 4.5 | GO:0048185 | activin binding(GO:0048185) |
| 0.6 | 6.3 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.6 | 2.5 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.6 | 1.8 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.6 | 12.6 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.6 | 5.3 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.6 | 2.2 | GO:0098882 | structural constituent of presynaptic active zone(GO:0098882) |
| 0.5 | 8.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.5 | 1.6 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.5 | 4.7 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.5 | 9.3 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.5 | 5.7 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.5 | 2.6 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.5 | 30.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.5 | 6.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.5 | 1.9 | GO:1990518 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) single-stranded DNA-dependent ATP-dependent 3'-5' DNA helicase activity(GO:1990518) |
| 0.5 | 2.3 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.5 | 14.4 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.5 | 1.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.4 | 1.7 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.4 | 6.0 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.4 | 3.0 | GO:0031682 | G-protein gamma-subunit binding(GO:0031682) |
| 0.4 | 4.6 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.4 | 7.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.4 | 1.6 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.4 | 2.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.4 | 4.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.4 | 2.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.4 | 3.6 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.4 | 6.8 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.4 | 5.3 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.3 | 6.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.3 | 3.4 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.3 | 4.7 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.3 | 12.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.3 | 1.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.3 | 18.4 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.3 | 1.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.3 | 1.6 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.3 | 23.6 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.3 | 1.9 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.3 | 0.9 | GO:0016918 | retinal binding(GO:0016918) |
| 0.3 | 2.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 0.9 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.3 | 22.9 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.3 | 1.2 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.3 | 2.0 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.3 | 1.5 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.3 | 5.8 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.3 | 10.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.3 | 1.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.3 | 2.0 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.3 | 1.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.3 | 10.6 | GO:0019209 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.3 | 7.8 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.3 | 0.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.3 | 5.1 | GO:0019211 | phosphatase activator activity(GO:0019211) protein phosphatase activator activity(GO:0072542) |
| 0.3 | 3.7 | GO:1990782 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.3 | 3.5 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.3 | 2.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.3 | 5.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.3 | 2.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.3 | 1.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.2 | 1.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 11.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.2 | 1.5 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.2 | 2.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 9.8 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 6.0 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.2 | 3.9 | GO:0019210 | protein kinase inhibitor activity(GO:0004860) kinase inhibitor activity(GO:0019210) |
| 0.2 | 2.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.2 | 2.9 | GO:0042562 | hormone binding(GO:0042562) |
| 0.2 | 2.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.2 | 2.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.2 | 1.0 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.2 | 12.4 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.2 | 1.6 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.2 | 90.1 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.2 | 1.2 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 0.2 | 3.1 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.2 | 1.0 | GO:0003707 | steroid hormone receptor activity(GO:0003707) |
| 0.2 | 2.0 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 2.1 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.2 | 4.5 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.2 | 7.6 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.2 | 4.4 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.2 | 3.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.2 | 0.7 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.2 | 3.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.2 | 0.7 | GO:0004422 | hypoxanthine phosphoribosyltransferase activity(GO:0004422) |
| 0.2 | 0.7 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 2.6 | GO:0061608 | nuclear import signal receptor activity(GO:0061608) |
| 0.2 | 2.7 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 6.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 10.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 2.4 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 1.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.1 | 1.3 | GO:0032977 | membrane insertase activity(GO:0032977) |
| 0.1 | 0.7 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 2.4 | GO:0098631 | protein binding involved in cell adhesion(GO:0098631) |
| 0.1 | 10.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.1 | 0.7 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.5 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 5.8 | GO:0019212 | phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 5.6 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.1 | 2.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 1.2 | GO:0016208 | AMP binding(GO:0016208) |
| 0.1 | 0.6 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.1 | 1.7 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 1.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.5 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 3.6 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 4.1 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 2.1 | GO:0033764 | steroid dehydrogenase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0033764) |
| 0.1 | 1.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 0.3 | GO:0003874 | 6-pyruvoyltetrahydropterin synthase activity(GO:0003874) |
| 0.1 | 1.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.1 | 4.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 2.6 | GO:0008528 | G-protein coupled peptide receptor activity(GO:0008528) |
| 0.1 | 0.3 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.1 | 0.3 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 1.5 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 3.6 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.1 | 2.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.6 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.1 | 0.5 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.3 | GO:0004485 | methylcrotonoyl-CoA carboxylase activity(GO:0004485) |
| 0.1 | 0.4 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.1 | 5.1 | GO:0050660 | flavin adenine dinucleotide binding(GO:0050660) |
| 0.1 | 3.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.1 | 24.1 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 3.4 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.1 | 7.5 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.1 | 1.8 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 1.7 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.1 | 3.0 | GO:0051287 | NAD binding(GO:0051287) |
| 0.1 | 0.8 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.1 | 7.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.1 | 2.2 | GO:0015171 | amino acid transmembrane transporter activity(GO:0015171) |
| 0.1 | 9.0 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.1 | 1.6 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.8 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 2.0 | GO:0045499 | semaphorin receptor binding(GO:0030215) chemorepellent activity(GO:0045499) |
| 0.1 | 0.9 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 9.5 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 0.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 51.0 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 0.8 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 3.2 | GO:0042626 | hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances(GO:0016820) ATPase activity, coupled to transmembrane movement of substances(GO:0042626) ATPase activity, coupled to movement of substances(GO:0043492) |
| 0.0 | 2.0 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 1.7 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.7 | GO:0019955 | cytokine binding(GO:0019955) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.5 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.5 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.1 | GO:0035258 | steroid hormone receptor binding(GO:0035258) |
| 0.0 | 0.6 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 1.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 5.2 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.3 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 9.9 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.2 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.0 | 1.3 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.2 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 10.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.2 | GO:0004724 | magnesium-dependent protein serine/threonine phosphatase activity(GO:0004724) |
| 0.0 | 2.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.4 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.0 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.3 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 20.9 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.9 | 2.8 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.8 | 17.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.7 | 8.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.6 | 1.7 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.5 | 7.6 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.4 | 14.7 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.4 | 0.9 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.4 | 6.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.4 | 7.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.4 | 3.9 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.3 | 3.5 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 8.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.3 | 4.8 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.3 | 5.4 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.3 | 7.0 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.3 | 2.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.3 | 3.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.3 | 6.0 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.3 | 9.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.3 | 9.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.3 | 6.2 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.3 | 22.7 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.2 | 0.5 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.2 | 4.6 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.2 | 4.1 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.2 | 0.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.2 | 2.8 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 7.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.2 | 5.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.2 | 2.5 | PID EPO PATHWAY | EPO signaling pathway |
| 0.2 | 2.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 6.9 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.1 | 6.7 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.1 | 2.7 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.1 | 1.0 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 2.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 1.0 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.1 | 1.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 1.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.1 | 1.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 2.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 0.6 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.1 | 1.9 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 0.9 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 2.9 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.1 | 1.8 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.1 | 3.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.2 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 1.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.1 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 0.3 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 23.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 1.7 | 7.0 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 1.3 | 6.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 1.3 | 9.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 1.2 | 9.9 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 1.2 | 11.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.9 | 12.1 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.8 | 6.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.8 | 2.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.7 | 6.2 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.7 | 4.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.6 | 14.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.6 | 12.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.6 | 2.8 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.6 | 2.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.5 | 1.5 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.5 | 1.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.5 | 16.6 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.5 | 14.3 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.5 | 5.5 | REACTOME BOTULINUM NEUROTOXICITY | Genes involved in Botulinum neurotoxicity |
| 0.4 | 3.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.4 | 4.7 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.4 | 3.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.4 | 3.5 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.4 | 7.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.4 | 3.1 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.4 | 3.9 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.4 | 7.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.3 | 3.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.3 | 6.5 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.3 | 2.9 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.3 | 5.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 1.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.3 | 3.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.3 | 5.7 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.3 | 12.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.2 | 2.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.2 | 4.2 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.2 | 3.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 8.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.2 | 2.9 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.2 | 1.5 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.2 | 5.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.2 | 1.6 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.2 | 5.1 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.2 | 2.1 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.2 | 2.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.2 | 2.4 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.2 | 2.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 2.0 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.1 | 10.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.1 | 10.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 2.0 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.1 | 1.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.1 | 0.9 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.1 | 1.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.1 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.1 | 5.9 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 2.9 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.1 | 0.6 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.1 | 2.5 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 2.5 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.1 | 1.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 2.5 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.1 | 1.9 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.1 | 1.0 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 1.5 | REACTOME TRANSLATION | Genes involved in Translation |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.0 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.7 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 0.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 3.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 2.0 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.3 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.0 | 1.4 | REACTOME SIGNALING BY PDGF | Genes involved in Signaling by PDGF |
| 0.0 | 0.7 | REACTOME CHONDROITIN SULFATE DERMATAN SULFATE METABOLISM | Genes involved in Chondroitin sulfate/dermatan sulfate metabolism |
| 0.0 | 0.6 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |