Project
DANIO-CODE
Navigation
Downloads
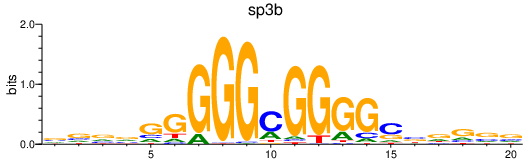
Results for sp3b
Z-value: 1.45
Transcription factors associated with sp3b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
sp3b
|
ENSDARG00000007812 | Sp3b transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| sp3b | dr10_dc_chr6_-_10492123_10492168 | 0.63 | 9.0e-03 | Click! |
Activity profile of sp3b motif
Sorted Z-values of sp3b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of sp3b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_44892852 | 4.43 |
ENSDART00000160961
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr14_+_268119 | 4.22 |
|
|
|
| chr7_+_44463158 | 4.15 |
ENSDART00000066380
|
ca7
|
carbonic anhydrase VII |
| KN149861v1_-_5655 | 4.09 |
|
|
|
| KN150589v1_-_5839 | 3.64 |
ENSDART00000157761
ENSDART00000157531 |
elovl7b
|
ELOVL fatty acid elongase 7b |
| chr23_-_31719203 | 3.60 |
ENSDART00000148122
|
sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr3_+_50398735 | 3.44 |
|
|
|
| chr8_-_410190 | 3.40 |
ENSDART00000151155
|
trim36
|
tripartite motif containing 36 |
| chr16_+_25344257 | 3.31 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr5_+_65453249 | 3.26 |
ENSDART00000073892
|
zgc:114041
|
zgc:114041 |
| chr6_+_3174047 | 3.26 |
ENSDART00000160290
|
st3gal3a
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 3a |
| chr17_-_53241446 | 3.12 |
|
|
|
| chr25_+_22222388 | 3.10 |
ENSDART00000154376
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr3_-_18653909 | 3.08 |
|
|
|
| chr1_-_44893257 | 2.99 |
ENSDART00000160961
|
atf7ip
|
activating transcription factor 7 interacting protein |
| chr20_-_194135 | 2.98 |
ENSDART00000064546
|
si:ch211-241j12.3
|
si:ch211-241j12.3 |
| chr13_+_969851 | 2.97 |
ENSDART00000139560
|
ppp3r1a
|
protein phosphatase 3, regulatory subunit B, alpha a |
| KN149861v1_-_6089 | 2.79 |
|
|
|
| KN149861v1_-_6661 | 2.76 |
|
|
|
| chr22_-_21021942 | 2.73 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| chr7_-_19116999 | 2.70 |
ENSDART00000165680
|
ntn4
|
netrin 4 |
| chr10_-_4641624 | 2.66 |
ENSDART00000163951
|
plppr1
|
phospholipid phosphatase related 1 |
| chr25_+_21001126 | 2.66 |
ENSDART00000079012
|
rad52
|
RAD52 homolog, DNA repair protein |
| chr25_+_16259634 | 2.64 |
ENSDART00000136454
|
tead1a
|
TEA domain family member 1a |
| chr8_-_26690715 | 2.63 |
ENSDART00000135215
|
tmem51a
|
transmembrane protein 51a |
| chr15_-_33669618 | 2.61 |
ENSDART00000161151
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr16_-_29452509 | 2.51 |
ENSDART00000148787
|
s100a1
|
S100 calcium binding protein A1 |
| chr22_+_866803 | 2.50 |
ENSDART00000140588
|
stk38b
|
serine/threonine kinase 38b |
| chr17_-_52233071 | 2.49 |
|
|
|
| chr24_+_41991604 | 2.46 |
|
|
|
| chr24_-_7534334 | 2.46 |
|
|
|
| chr23_+_19728953 | 2.43 |
ENSDART00000104441
|
abhd6b
|
abhydrolase domain containing 6b |
| KN150349v1_-_1437 | 2.42 |
|
|
|
| chr15_-_5913264 | 2.38 |
ENSDART00000155156
ENSDART00000155971 |
si:ch73-281n10.2
|
si:ch73-281n10.2 |
| chr7_-_73613531 | 2.38 |
ENSDART00000128137
|
zgc:92594
|
zgc:92594 |
| chr5_-_56641061 | 2.38 |
ENSDART00000050957
|
fer
|
fer (fps/fes related) tyrosine kinase |
| chr17_-_45021393 | 2.37 |
|
|
|
| chr22_-_111800 | 2.37 |
ENSDART00000163198
ENSDART00000168678 |
capzb
|
capping protein (actin filament) muscle Z-line, beta |
| chr18_-_26732762 | 2.37 |
ENSDART00000142043
|
malt3
|
MALT paracaspase 3 |
| chr5_-_56641212 | 2.36 |
ENSDART00000050957
|
fer
|
fer (fps/fes related) tyrosine kinase |
| chr16_-_39655528 | 2.35 |
ENSDART00000146301
|
tgfbr2b
|
transforming growth factor beta receptor 2b |
| chr15_+_25554119 | 2.34 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr2_-_44330306 | 2.34 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr20_-_50089784 | 2.32 |
ENSDART00000058858
|
kif13bb
|
kinesin family member 13Bb |
| chr22_-_11817641 | 2.32 |
ENSDART00000000192
|
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr20_+_37891739 | 2.31 |
ENSDART00000022060
|
atf3
|
activating transcription factor 3 |
| chr7_-_73890600 | 2.31 |
ENSDART00000164992
|
rbpms
|
RNA binding protein with multiple splicing |
| chr13_-_15662476 | 2.31 |
ENSDART00000145914
ENSDART00000010286 |
bag5
|
BCL2-associated athanogene 5 |
| chr6_+_41506350 | 2.29 |
ENSDART00000136538
|
cish
|
cytokine inducible SH2-containing protein |
| chr6_+_41506220 | 2.24 |
ENSDART00000136538
|
cish
|
cytokine inducible SH2-containing protein |
| chr22_-_11817587 | 2.24 |
ENSDART00000000192
|
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr19_+_14132473 | 2.24 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr23_+_25062876 | 2.23 |
ENSDART00000145307
ENSDART00000172299 |
arhgap4a
|
Rho GTPase activating protein 4a |
| chr7_-_61373495 | 2.19 |
ENSDART00000098622
|
lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr13_-_15662599 | 2.18 |
ENSDART00000145914
ENSDART00000010286 |
bag5
|
BCL2-associated athanogene 5 |
| chr7_-_73890499 | 2.15 |
ENSDART00000164992
|
rbpms
|
RNA binding protein with multiple splicing |
| chr19_+_14132374 | 2.15 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr8_-_3974320 | 2.13 |
ENSDART00000159895
|
mtmr3
|
myotubularin related protein 3 |
| chr7_-_64745132 | 2.12 |
ENSDART00000112166
|
fam60al
|
family with sequence similarity 60, member A, like |
| chr17_+_14957568 | 2.12 |
ENSDART00000066604
|
gpr137c
|
G protein-coupled receptor 137c |
| chr5_-_29480173 | 2.10 |
ENSDART00000140049
|
bco2a
|
beta-carotene oxygenase 2a |
| chr1_-_44907320 | 2.09 |
|
|
|
| chr7_-_19080002 | 2.08 |
ENSDART00000166121
ENSDART00000169668 |
dock11
|
dedicator of cytokinesis 11 |
| chr6_+_41506504 | 2.07 |
ENSDART00000084861
|
cish
|
cytokine inducible SH2-containing protein |
| chr16_-_34471672 | 2.06 |
ENSDART00000172162
|
CR626886.2
|
ENSDARG00000105308 |
| chr24_-_38197984 | 2.06 |
ENSDART00000078998
|
crp2
|
C-reactive protein 2 |
| chr7_+_71263100 | 2.04 |
ENSDART00000100396
|
ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr7_+_13742576 | 2.03 |
|
|
|
| KN150589v1_-_5785 | 2.02 |
ENSDART00000157761
ENSDART00000157531 |
elovl7b
|
ELOVL fatty acid elongase 7b |
| chr19_+_26756510 | 2.01 |
ENSDART00000013497
|
mylipa
|
myosin regulatory light chain interacting protein a |
| KN149861v1_-_5543 | 2.01 |
|
|
|
| chr19_+_14490203 | 2.00 |
ENSDART00000164386
|
arid1ab
|
AT rich interactive domain 1Ab (SWI-like) |
| chr23_-_14118530 | 2.00 |
ENSDART00000139406
|
g6pd
|
glucose-6-phosphate dehydrogenase |
| chr18_+_46153484 | 1.99 |
ENSDART00000015034
|
blvrb
|
biliverdin reductase B |
| chr5_-_22466262 | 1.99 |
ENSDART00000172549
|
si:dkey-237j10.2
|
si:dkey-237j10.2 |
| chr22_-_21873054 | 1.98 |
|
|
|
| chr3_+_51430252 | 1.98 |
ENSDART00000159493
|
baiap2a
|
BAI1-associated protein 2a |
| chr3_-_58400454 | 1.98 |
ENSDART00000052179
|
cdr2a
|
cerebellar degeneration-related protein 2a |
| chr15_+_29092022 | 1.98 |
ENSDART00000141164
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr22_+_26840517 | 1.97 |
ENSDART00000158756
|
crebbpa
|
CREB binding protein a |
| chr8_-_410293 | 1.95 |
ENSDART00000164886
|
trim36
|
tripartite motif containing 36 |
| chr7_+_1443102 | 1.94 |
ENSDART00000172830
|
supt16h
|
SPT16 homolog, facilitates chromatin remodeling subunit |
| chr2_+_44659334 | 1.94 |
ENSDART00000155017
|
pask
|
PAS domain containing serine/threonine kinase |
| chr22_-_35003371 | 1.91 |
ENSDART00000133537
|
arhgap19
|
Rho GTPase activating protein 19 |
| chr22_-_10722533 | 1.90 |
ENSDART00000081191
|
sass6
|
SAS-6 centriolar assembly protein |
| chr17_+_30476515 | 1.87 |
|
|
|
| chr17_+_11921705 | 1.87 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr12_+_46641510 | 1.87 |
ENSDART00000008009
|
trim16
|
tripartite motif containing 16 |
| chr15_-_33669589 | 1.86 |
ENSDART00000161151
|
stard13b
|
StAR-related lipid transfer (START) domain containing 13b |
| chr19_-_5142209 | 1.84 |
ENSDART00000150980
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| KN149861v1_-_6916 | 1.81 |
ENSDART00000169003
|
5_8S_rRNA
|
5.8S ribosomal RNA |
| chr21_+_10485148 | 1.81 |
ENSDART00000165070
|
ccbe1
|
collagen and calcium binding EGF domains 1 |
| chr23_+_35327492 | 1.80 |
ENSDART00000164658
|
si:ch211-225h24.2
|
si:ch211-225h24.2 |
| chr6_+_41506446 | 1.80 |
ENSDART00000140108
|
cish
|
cytokine inducible SH2-containing protein |
| chr16_-_52847564 | 1.80 |
ENSDART00000147236
|
azin1a
|
antizyme inhibitor 1a |
| chr13_-_17729510 | 1.80 |
ENSDART00000170583
|
march8
|
membrane-associated ring finger (C3HC4) 8 |
| chr2_-_52956254 | 1.78 |
ENSDART00000044411
|
gna11b
|
guanine nucleotide binding protein (G protein), alpha 11b (Gq class) |
| chr7_-_73613763 | 1.78 |
ENSDART00000128137
|
zgc:92594
|
zgc:92594 |
| chr11_+_33154781 | 1.77 |
|
|
|
| chr16_+_25344184 | 1.77 |
ENSDART00000058938
|
fbxo32
|
F-box protein 32 |
| chr7_-_19080041 | 1.77 |
ENSDART00000137575
|
dock11
|
dedicator of cytokinesis 11 |
| chr23_-_42918086 | 1.77 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr19_-_6274169 | 1.75 |
ENSDART00000140347
ENSDART00000092656 |
erf
|
Ets2 repressor factor |
| chr5_-_56641001 | 1.73 |
ENSDART00000149855
|
fer
|
fer (fps/fes related) tyrosine kinase |
| chr15_-_18087085 | 1.73 |
ENSDART00000155194
|
phldb1b
|
pleckstrin homology-like domain, family B, member 1b |
| chr17_+_12788541 | 1.73 |
ENSDART00000016597
|
nfkbiab
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha b |
| chr25_+_22222336 | 1.72 |
ENSDART00000154065
|
cyp11a1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr20_+_4034072 | 1.72 |
ENSDART00000092217
|
ttc13
|
tetratricopeptide repeat domain 13 |
| chr19_-_7402329 | 1.72 |
ENSDART00000092375
|
oxr1b
|
oxidation resistance 1b |
| chr23_+_25062826 | 1.71 |
ENSDART00000172299
|
arhgap4a
|
Rho GTPase activating protein 4a |
| chr22_-_20377970 | 1.70 |
ENSDART00000010048
|
map2k2a
|
mitogen-activated protein kinase kinase 2a |
| chr17_+_11921923 | 1.69 |
ENSDART00000155329
|
cnsta
|
consortin, connexin sorting protein a |
| chr13_-_44671682 | 1.68 |
ENSDART00000099984
|
glo1
|
glyoxalase 1 |
| chr1_+_23778481 | 1.68 |
ENSDART00000176581
|
fam160a1a
|
family with sequence similarity 160, member A1a |
| chr20_+_37891795 | 1.68 |
ENSDART00000022060
|
atf3
|
activating transcription factor 3 |
| chr10_-_625501 | 1.65 |
ENSDART00000041236
ENSDART00000144882 |
rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr3_-_9755642 | 1.65 |
ENSDART00000168366
|
crebbpb
|
CREB binding protein b |
| chr2_-_52956344 | 1.64 |
ENSDART00000044411
|
gna11b
|
guanine nucleotide binding protein (G protein), alpha 11b (Gq class) |
| chr14_+_11803542 | 1.64 |
ENSDART00000121913
|
kctd12b
|
potassium channel tetramerisation domain containing 12b |
| chr3_+_26682186 | 1.63 |
ENSDART00000055537
|
socs1a
|
suppressor of cytokine signaling 1a |
| chr7_+_71262845 | 1.63 |
ENSDART00000100396
|
ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr25_+_4508802 | 1.60 |
ENSDART00000021120
|
pgghg
|
protein-glucosylgalactosylhydroxylysine glucosidase |
| chr19_+_1298795 | 1.60 |
ENSDART00000113368
|
RNF5
|
ring finger protein 5 |
| chr24_+_39339095 | 1.59 |
ENSDART00000168705
|
si:ch73-103b11.2
|
si:ch73-103b11.2 |
| chr8_-_26690680 | 1.58 |
ENSDART00000135215
|
tmem51a
|
transmembrane protein 51a |
| chr13_-_31267133 | 1.58 |
ENSDART00000008287
|
pgam1a
|
phosphoglycerate mutase 1a |
| chr7_-_61373339 | 1.57 |
ENSDART00000148270
|
lrch4
|
leucine-rich repeats and calponin homology (CH) domain containing 4 |
| chr20_+_16274045 | 1.56 |
ENSDART00000129633
|
zyg11
|
zyg-11 homolog (C. elegans) |
| chr1_+_13768098 | 1.56 |
ENSDART00000005067
|
rbpja
|
recombination signal binding protein for immunoglobulin kappa J region a |
| chr19_-_5142238 | 1.56 |
ENSDART00000130062
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr23_+_35327654 | 1.56 |
ENSDART00000165034
|
si:ch211-225h24.2
|
si:ch211-225h24.2 |
| chr14_+_25950244 | 1.55 |
ENSDART00000113804
ENSDART00000159054 |
CCDC69
|
coiled-coil domain containing 69 |
| chr15_-_30935719 | 1.55 |
ENSDART00000050649
|
msi2b
|
musashi RNA-binding protein 2b |
| chr7_-_45579815 | 1.54 |
ENSDART00000170224
|
shcbp1
|
SHC SH2-domain binding protein 1 |
| chr4_-_17364090 | 1.54 |
ENSDART00000134467
|
parpbp
|
PARP1 binding protein |
| chr10_+_384571 | 1.54 |
ENSDART00000129757
|
si:ch211-242f23.8
|
si:ch211-242f23.8 |
| chr3_+_29338315 | 1.54 |
ENSDART00000103592
|
fam83fa
|
family with sequence similarity 83, member Fa |
| chr16_-_39655605 | 1.54 |
ENSDART00000146301
|
tgfbr2b
|
transforming growth factor beta receptor 2b |
| chr15_+_11760631 | 1.52 |
|
|
|
| chr2_-_52956156 | 1.51 |
ENSDART00000044411
|
gna11b
|
guanine nucleotide binding protein (G protein), alpha 11b (Gq class) |
| chr19_-_7402373 | 1.51 |
ENSDART00000092375
|
oxr1b
|
oxidation resistance 1b |
| chr15_-_43283562 | 1.51 |
ENSDART00000110352
|
ap1s3a
|
adaptor-related protein complex 1, sigma 3 subunit, a |
| chr15_-_17088654 | 1.50 |
ENSDART00000154719
ENSDART00000014465 |
hip1
|
huntingtin interacting protein 1 |
| chr6_+_28001318 | 1.50 |
ENSDART00000143974
|
amotl2a
|
angiomotin like 2a |
| chr2_+_12547264 | 1.49 |
ENSDART00000169724
|
zgc:66475
|
zgc:66475 |
| chr2_-_47766563 | 1.48 |
ENSDART00000038228
|
ap1s3b
|
adaptor-related protein complex 1, sigma 3 subunit, b |
| chr23_+_39454172 | 1.48 |
ENSDART00000149819
|
src
|
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
| chr20_-_14885722 | 1.48 |
ENSDART00000160481
|
suco
|
SUN domain containing ossification factor |
| chr4_-_4603205 | 1.47 |
ENSDART00000130601
|
CABZ01020840.1
|
ENSDARG00000090401 |
| chr3_-_58400345 | 1.47 |
ENSDART00000052179
|
cdr2a
|
cerebellar degeneration-related protein 2a |
| chr16_-_34471475 | 1.47 |
ENSDART00000172162
|
CR626886.2
|
ENSDARG00000105308 |
| chr24_-_38197866 | 1.46 |
ENSDART00000078998
|
crp2
|
C-reactive protein 2 |
| chr6_-_10574082 | 1.45 |
ENSDART00000151661
|
wipf1b
|
WAS/WASL interacting protein family, member 1b |
| chr5_-_6054581 | 1.45 |
|
|
|
| chr22_-_21021984 | 1.45 |
ENSDART00000133982
|
ssbp4
|
single stranded DNA binding protein 4 |
| KN150593v1_+_10332 | 1.44 |
|
|
|
| chr2_+_35621136 | 1.44 |
ENSDART00000133018
ENSDART00000147278 |
plk3
|
polo-like kinase 3 (Drosophila) |
| chr23_-_42918055 | 1.43 |
ENSDART00000102328
|
pfkfb2a
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2a |
| chr3_-_36298676 | 1.43 |
ENSDART00000157950
|
rogdi
|
rogdi homolog (Drosophila) |
| chr22_-_5888816 | 1.43 |
ENSDART00000141373
|
si:rp71-36a1.1
|
si:rp71-36a1.1 |
| chr1_+_28936288 | 1.42 |
|
|
|
| chr13_+_291022 | 1.42 |
ENSDART00000017854
|
lgi1a
|
leucine-rich, glioma inactivated 1a |
| chr17_+_25314229 | 1.42 |
ENSDART00000082319
|
tmem54a
|
transmembrane protein 54a |
| chr10_-_17214368 | 1.42 |
|
|
|
| chr22_+_818795 | 1.42 |
ENSDART00000003325
|
dennd2db
|
DENN/MADD domain containing 2Db |
| chr15_+_40340914 | 1.41 |
ENSDART00000063779
|
efhd1
|
EF-hand domain family, member D1 |
| chr13_-_22776767 | 1.40 |
ENSDART00000143097
|
rufy2
|
RUN and FYVE domain containing 2 |
| chr2_+_25212281 | 1.37 |
ENSDART00000078838
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr14_+_46609215 | 1.37 |
ENSDART00000110429
|
zgc:194285
|
zgc:194285 |
| chr13_-_37043725 | 1.37 |
|
|
|
| chr5_-_1102450 | 1.37 |
ENSDART00000115044
|
mbd2
|
methyl-CpG binding domain protein 2 |
| chr16_-_34471544 | 1.37 |
ENSDART00000172162
|
CR626886.2
|
ENSDARG00000105308 |
| chr19_-_44161951 | 1.37 |
ENSDART00000058491
|
ppt1
|
palmitoyl-protein thioesterase 1 (ceroid-lipofuscinosis, neuronal 1, infantile) |
| chr24_+_41991566 | 1.37 |
|
|
|
| chr12_-_37274632 | 1.36 |
ENSDART00000152951
|
cdc42ep4b
|
CDC42 effector protein (Rho GTPase binding) 4b |
| chr13_-_50053501 | 1.36 |
ENSDART00000098209
|
sirt1
|
sirtuin 1 |
| chr23_-_43785497 | 1.36 |
ENSDART00000165963
|
ENSDARG00000102050
|
ENSDARG00000102050 |
| chr20_-_54160297 | 1.35 |
ENSDART00000153435
|
ppp2r5cb
|
protein phosphatase 2, regulatory subunit B', gamma b |
| chr23_+_27777325 | 1.35 |
ENSDART00000134008
|
lmbr1l
|
limb development membrane protein 1-like |
| chr24_+_39339391 | 1.35 |
ENSDART00000168705
|
si:ch73-103b11.2
|
si:ch73-103b11.2 |
| chr22_-_20899539 | 1.33 |
ENSDART00000100642
|
ell
|
elongation factor RNA polymerase II |
| chr15_+_29091983 | 1.33 |
ENSDART00000141164
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr7_+_13742622 | 1.33 |
|
|
|
| KN149704v1_-_17031 | 1.32 |
|
|
|
| chr17_-_53241366 | 1.31 |
ENSDART00000171082
|
exd1
|
exonuclease 3'-5' domain containing 1 |
| chr2_-_52955900 | 1.30 |
ENSDART00000044411
|
gna11b
|
guanine nucleotide binding protein (G protein), alpha 11b (Gq class) |
| chr24_+_37805069 | 1.30 |
|
|
|
| KN150286v1_-_2942 | 1.30 |
|
|
|
| chr20_+_46837436 | 1.29 |
ENSDART00000145294
|
ENSDARG00000022260
|
ENSDARG00000022260 |
| chr10_-_5023980 | 1.27 |
ENSDART00000137853
|
hnrpdl
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr5_+_60677040 | 1.27 |
ENSDART00000009507
|
gatsl2
|
GATS protein-like 2 |
| chr8_+_23144448 | 1.27 |
ENSDART00000063057
|
dnajc5aa
|
DnaJ (Hsp40) homolog, subfamily C, member 5aa |
| chr6_+_12268892 | 1.26 |
ENSDART00000171460
|
stk24b
|
serine/threonine kinase 24b (STE20 homolog, yeast) |
| chr11_-_40239946 | 1.25 |
ENSDART00000165394
|
si:dkeyp-61b2.1
|
si:dkeyp-61b2.1 |
| chr18_+_6498590 | 1.24 |
ENSDART00000162398
|
c2cd5
|
C2 calcium-dependent domain containing 5 |
| chr2_-_44330348 | 1.24 |
ENSDART00000111246
|
cadm3
|
cell adhesion molecule 3 |
| chr21_-_1339525 | 1.23 |
|
|
|
| chr14_+_25950198 | 1.22 |
ENSDART00000113804
ENSDART00000159054 |
CCDC69
|
coiled-coil domain containing 69 |
| chr16_+_54466704 | 1.22 |
ENSDART00000177364
ENSDART00000176635 |
CABZ01118317.1
|
ENSDARG00000108514 |
| chr19_-_27185558 | 1.22 |
ENSDART00000043776
ENSDART00000168777 ENSDART00000159489 |
prrc2a
|
proline-rich coiled-coil 2A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0006700 | C21-steroid hormone biosynthetic process(GO:0006700) |
| 1.5 | 4.5 | GO:0090083 | inclusion body assembly(GO:0070841) regulation of inclusion body assembly(GO:0090083) |
| 1.3 | 6.5 | GO:0038095 | Fc-epsilon receptor signaling pathway(GO:0038095) |
| 1.3 | 10.0 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 1.2 | 6.2 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 1.0 | 3.0 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.9 | 3.6 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.8 | 4.8 | GO:0045002 | DNA recombinase assembly(GO:0000730) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.8 | 3.9 | GO:0032516 | positive regulation of phosphatase activity(GO:0010922) positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.7 | 8.5 | GO:0090309 | methylation-dependent chromatin silencing(GO:0006346) positive regulation of chromatin silencing(GO:0031937) regulation of methylation-dependent chromatin silencing(GO:0090308) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.7 | 2.1 | GO:0016119 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.7 | 2.0 | GO:0098751 | bone cell development(GO:0098751) |
| 0.6 | 1.8 | GO:0060855 | venous endothelial cell migration involved in lymph vessel development(GO:0060855) |
| 0.5 | 2.9 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.5 | 1.4 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.4 | 6.1 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.4 | 1.3 | GO:0042754 | negative regulation of circadian rhythm(GO:0042754) |
| 0.4 | 1.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.4 | 1.6 | GO:0072149 | glomerular visceral epithelial cell fate commitment(GO:0072149) glomerular epithelial cell fate commitment(GO:0072314) |
| 0.4 | 2.3 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.4 | 1.9 | GO:0070874 | negative regulation of glycogen biosynthetic process(GO:0045719) negative regulation of glycogen metabolic process(GO:0070874) |
| 0.4 | 1.1 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.4 | 1.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.3 | 1.4 | GO:0048790 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.3 | 3.4 | GO:0060876 | semicircular canal formation(GO:0060876) |
| 0.3 | 2.7 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) regulation of lamellipodium organization(GO:1902743) |
| 0.3 | 1.3 | GO:0019075 | virion assembly(GO:0019068) virus maturation(GO:0019075) multi-organism membrane organization(GO:0044803) viral budding(GO:0046755) multi-organism organelle organization(GO:1902590) multi-organism membrane budding(GO:1902592) |
| 0.3 | 1.5 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.3 | 0.9 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.3 | 2.4 | GO:0046850 | regulation of bone remodeling(GO:0046850) |
| 0.3 | 1.2 | GO:0070940 | termination of RNA polymerase II transcription(GO:0006369) dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.3 | 4.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.3 | 2.6 | GO:0035912 | aorta morphogenesis(GO:0035909) dorsal aorta morphogenesis(GO:0035912) |
| 0.3 | 1.3 | GO:0042795 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.3 | 0.8 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.2 | 2.0 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.2 | 2.9 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.2 | 2.1 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.2 | 0.7 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.2 | 1.5 | GO:0021772 | olfactory bulb development(GO:0021772) olfactory lobe development(GO:0021988) |
| 0.2 | 1.3 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 5.7 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.2 | 1.8 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.2 | 6.1 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.2 | 1.1 | GO:0032366 | intracellular sterol transport(GO:0032366) |
| 0.2 | 0.9 | GO:0006561 | proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.2 | 2.0 | GO:0031269 | pseudopodium organization(GO:0031268) pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.1 | 0.9 | GO:0044857 | membrane raft assembly(GO:0001765) plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.1 | 1.9 | GO:0034724 | DNA replication-independent nucleosome organization(GO:0034724) |
| 0.1 | 0.7 | GO:1904325 | inhibitory G-protein coupled receptor phosphorylation(GO:0002030) positive regulation of calcium ion transport into cytosol(GO:0010524) positive regulation of release of sequestered calcium ion into cytosol(GO:0051281) regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904323) positive regulation of inhibitory G-protein coupled receptor phosphorylation(GO:1904325) |
| 0.1 | 0.9 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.1 | 1.4 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.6 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.1 | 0.8 | GO:0032534 | regulation of microvillus organization(GO:0032530) regulation of microvillus assembly(GO:0032534) |
| 0.1 | 1.7 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.1 | 1.7 | GO:0050796 | cilium or flagellum-dependent cell motility(GO:0001539) regulation of insulin secretion(GO:0050796) cilium-dependent cell motility(GO:0060285) |
| 0.1 | 0.8 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.6 | GO:0003128 | heart field specification(GO:0003128) |
| 0.1 | 0.3 | GO:0046661 | development of primary male sexual characteristics(GO:0046546) male sex differentiation(GO:0046661) |
| 0.1 | 1.4 | GO:0002084 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.4 | GO:0010990 | SMAD protein complex assembly(GO:0007183) regulation of SMAD protein complex assembly(GO:0010990) negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 1.2 | GO:0097499 | protein localization to nonmotile primary cilium(GO:0097499) |
| 0.1 | 1.5 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.3 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.1 | 0.3 | GO:0071364 | response to epidermal growth factor(GO:0070849) cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.1 | 4.0 | GO:0009615 | response to virus(GO:0009615) |
| 0.1 | 0.9 | GO:0099601 | regulation of neurotransmitter receptor activity(GO:0099601) |
| 0.1 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 4.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.1 | 0.5 | GO:0050805 | negative regulation of synaptic transmission(GO:0050805) negative regulation of neurotransmitter transport(GO:0051589) |
| 0.1 | 1.2 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.8 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 3.9 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.1 | 0.9 | GO:0042026 | protein refolding(GO:0042026) |
| 0.1 | 0.9 | GO:0060998 | regulation of dendritic spine development(GO:0060998) |
| 0.1 | 0.9 | GO:0048016 | calcineurin-NFAT signaling cascade(GO:0033173) inositol phosphate-mediated signaling(GO:0048016) |
| 0.1 | 0.4 | GO:0007063 | regulation of sister chromatid cohesion(GO:0007063) regulation of cohesin loading(GO:0071922) |
| 0.1 | 0.8 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.1 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 1.0 | GO:0090481 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.1 | 2.2 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.1 | 4.2 | GO:0007179 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.1 | 0.5 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.1 | 0.6 | GO:0042430 | indole-containing compound metabolic process(GO:0042430) phenol-containing compound biosynthetic process(GO:0046189) |
| 0.1 | 3.5 | GO:0009617 | response to bacterium(GO:0009617) |
| 0.1 | 0.7 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.1 | 2.0 | GO:0030048 | actin filament-based movement(GO:0030048) |
| 0.1 | 0.9 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 0.5 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.1 | 1.4 | GO:0007131 | reciprocal meiotic recombination(GO:0007131) |
| 0.0 | 3.4 | GO:0007283 | spermatogenesis(GO:0007283) male gamete generation(GO:0048232) |
| 0.0 | 2.5 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 4.0 | GO:0006954 | inflammatory response(GO:0006954) |
| 0.0 | 0.5 | GO:0071786 | endoplasmic reticulum tubular network organization(GO:0071786) |
| 0.0 | 0.8 | GO:0030500 | regulation of bone mineralization(GO:0030500) regulation of biomineral tissue development(GO:0070167) |
| 0.0 | 0.2 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 2.0 | GO:0043044 | ATP-dependent chromatin remodeling(GO:0043044) |
| 0.0 | 1.0 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 4.0 | GO:0016573 | histone acetylation(GO:0016573) |
| 0.0 | 2.1 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.9 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.2 | GO:0045950 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.3 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.0 | 0.6 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.0 | 1.9 | GO:0035023 | regulation of Rho protein signal transduction(GO:0035023) |
| 0.0 | 0.3 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 2.9 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 1.0 | GO:0021591 | ventricular system development(GO:0021591) |
| 0.0 | 0.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.6 | GO:0071542 | dopaminergic neuron differentiation(GO:0071542) |
| 0.0 | 1.7 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.6 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 1.7 | GO:0042552 | ensheathment of neurons(GO:0007272) axon ensheathment(GO:0008366) myelination(GO:0042552) |
| 0.0 | 0.3 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 0.0 | 0.4 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 1.0 | GO:0051091 | positive regulation of sequence-specific DNA binding transcription factor activity(GO:0051091) |
| 0.0 | 0.3 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0061588 | calcium activated phospholipid scrambling(GO:0061588) |
| 0.0 | 0.8 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 1.4 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.5 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 2.1 | GO:0030178 | negative regulation of Wnt signaling pathway(GO:0030178) |
| 0.0 | 0.8 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.6 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 1.3 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.5 | GO:0030968 | endoplasmic reticulum unfolded protein response(GO:0030968) |
| 0.0 | 0.2 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.3 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 0.5 | GO:0051051 | negative regulation of transport(GO:0051051) |
| 0.0 | 0.6 | GO:0070121 | Kupffer's vesicle development(GO:0070121) |
| 0.0 | 0.7 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0098536 | deuterosome(GO:0098536) |
| 0.6 | 1.8 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.5 | 6.2 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.4 | 1.4 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.3 | 1.7 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.3 | 1.6 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.3 | 0.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.3 | 1.9 | GO:0035101 | FACT complex(GO:0035101) |
| 0.3 | 2.9 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.2 | 4.7 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 2.5 | GO:0035060 | brahma complex(GO:0035060) |
| 0.2 | 3.9 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.2 | 10.0 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 13.5 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.1 | 1.7 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 1.6 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 1.1 | GO:0098827 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 1.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 4.0 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 7.1 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 1.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.8 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 3.1 | GO:0030175 | filopodium(GO:0030175) |
| 0.1 | 2.2 | GO:0005844 | polysome(GO:0005844) |
| 0.1 | 0.3 | GO:0071065 | alpha9-beta1 integrin-vascular cell adhesion molecule-1 complex(GO:0071065) |
| 0.1 | 0.8 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 4.9 | GO:0045121 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.1 | 0.3 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.1 | 0.6 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 1.3 | GO:0043186 | P granule(GO:0043186) |
| 0.1 | 0.6 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.1 | 0.3 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 4.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.3 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 4.0 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 5.7 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 3.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.4 | GO:0098978 | glutamatergic synapse(GO:0098978) |
| 0.0 | 2.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.3 | GO:0089701 | U2AF(GO:0089701) |
| 0.0 | 2.8 | GO:0048475 | membrane coat(GO:0030117) coated membrane(GO:0048475) |
| 0.0 | 1.8 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 2.1 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 2.9 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.1 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.2 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 1.2 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 1.3 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.0 | 1.8 | GO:0098852 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.4 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.9 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.3 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 1.4 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 0.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.6 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.3 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.8 | GO:0016713 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 1.3 | 3.9 | GO:0034713 | transforming growth factor beta receptor activity, type II(GO:0005026) type I transforming growth factor beta receptor binding(GO:0034713) |
| 1.1 | 3.3 | GO:0008118 | N-acetyllactosaminide alpha-2,3-sialyltransferase activity(GO:0008118) |
| 1.0 | 3.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.8 | 7.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.7 | 2.1 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.6 | 1.8 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.5 | 3.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.5 | 6.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.3 | 10.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.3 | 4.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.3 | 4.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.3 | 0.9 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.3 | 1.6 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.2 | 1.4 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.2 | 4.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 1.7 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.2 | 1.3 | GO:0015385 | monovalent cation:proton antiporter activity(GO:0005451) sodium:proton antiporter activity(GO:0015385) potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 5.7 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.2 | 3.4 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 0.6 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 0.8 | GO:0004630 | phospholipase D activity(GO:0004630) |
| 0.2 | 0.8 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.2 | 3.6 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.2 | 1.5 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.2 | 0.5 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.2 | 2.5 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.2 | 0.8 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.2 | 1.4 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.1 | 2.8 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 2.0 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.1 | 0.6 | GO:0051500 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 1.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.1 | 0.7 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 1.4 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 3.4 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 0.7 | GO:0060182 | apelin receptor activity(GO:0060182) |
| 0.1 | 0.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 0.6 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 0.3 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.1 | 0.6 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 0.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 1.6 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.1 | 3.7 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.1 | 1.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 1.0 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.3 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.1 | 4.0 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.1 | 1.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 1.1 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.3 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.1 | 5.8 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.1 | 1.4 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.3 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.1 | 0.8 | GO:0001130 | bacterial-type RNA polymerase transcription factor activity, sequence-specific DNA binding(GO:0001130) bacterial-type RNA polymerase transcriptional repressor activity, sequence-specific DNA binding(GO:0001217) |
| 0.1 | 1.4 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.1 | 0.3 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.1 | 1.1 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.1 | 0.3 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 1.7 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.1 | 0.2 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.7 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.9 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.8 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 1.0 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 2.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.4 | GO:0044390 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.5 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.7 | GO:0034061 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.0 | 4.1 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 0.3 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 3.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 2.1 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.8 | GO:0070851 | growth factor receptor binding(GO:0070851) |
| 0.0 | 1.3 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.5 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 8.1 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 3.9 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 3.1 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.5 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 8.4 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.5 | 2.0 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.2 | 11.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.2 | 6.5 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 0.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 3.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 3.8 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.1 | 3.7 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.1 | 1.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
| 0.1 | 3.4 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.8 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 3.9 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.5 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.2 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.0 | 0.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.5 | 4.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.4 | 4.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.4 | 8.4 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.2 | 4.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.2 | 2.6 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.2 | 3.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.2 | 0.5 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.2 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 3.9 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.1 | 3.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.1 | 5.9 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.1 | 1.1 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 0.7 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.1 | 3.1 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.1 | 0.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 0.6 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 0.8 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.4 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 8.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.0 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 1.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.7 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |