Project
DANIO-CODE
Navigation
Downloads
Results for spi2
Z-value: 2.48
Transcription factors associated with spi2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
spi2
|
ENSDARG00000087438 | Spi-2 proto-oncogene |
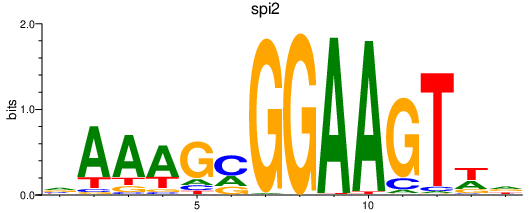
Activity profile of spi2 motif
Sorted Z-values of spi2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of spi2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_46426385 | 6.70 |
ENSDART00000141331
|
si:ch73-59c19.1
|
si:ch73-59c19.1 |
| chr2_-_38305602 | 6.44 |
ENSDART00000061677
|
si:ch211-14a17.6
|
si:ch211-14a17.6 |
| chr12_+_13053552 | 5.86 |
ENSDART00000124799
|
si:ch211-103b1.2
|
si:ch211-103b1.2 |
| chr9_+_8429700 | 5.24 |
ENSDART00000144373
|
zgc:153499
|
zgc:153499 |
| chr23_+_25062876 | 4.96 |
ENSDART00000145307
ENSDART00000172299 |
arhgap4a
|
Rho GTPase activating protein 4a |
| chr2_+_30199024 | 4.88 |
ENSDART00000004903
|
rdh10b
|
retinol dehydrogenase 10b |
| chr5_-_15971338 | 4.78 |
ENSDART00000110437
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr13_-_18564182 | 4.71 |
ENSDART00000176809
|
sfxn3
|
sideroflexin 3 |
| chr19_-_11486056 | 4.66 |
ENSDART00000109889
ENSDART00000172065 |
hsbp1l1
|
heat shock factor binding protein 1 like 1 |
| chr13_+_22973764 | 4.48 |
ENSDART00000110266
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr15_+_35081979 | 4.29 |
ENSDART00000131182
|
zgc:66024
|
zgc:66024 |
| chr18_+_284210 | 4.26 |
ENSDART00000158040
|
larp6
|
La ribonucleoprotein domain family, member 6 |
| chr11_-_2343333 | 4.02 |
ENSDART00000167032
ENSDART00000129854 |
tp53rk
|
TP53 regulating kinase |
| chr13_+_31586338 | 3.89 |
ENSDART00000034745
|
prkcha
|
protein kinase C, eta, a |
| chr23_+_25062826 | 3.88 |
ENSDART00000172299
|
arhgap4a
|
Rho GTPase activating protein 4a |
| chr20_-_46563338 | 3.81 |
ENSDART00000060702
|
rmdn3
|
regulator of microtubule dynamics 3 |
| chr16_-_21128542 | 3.79 |
ENSDART00000154833
|
CR392024.1
|
ENSDARG00000096565 |
| chr2_-_38329953 | 3.75 |
ENSDART00000159031
|
ap1g2
|
adaptor-related protein complex 1, gamma 2 subunit |
| chr4_-_3340315 | 3.53 |
ENSDART00000009076
|
pik3cg
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
| chr22_+_24596299 | 3.53 |
ENSDART00000158303
ENSDART00000160924 |
mcoln2
|
mucolipin 2 |
| chr16_-_41537827 | 3.51 |
ENSDART00000169312
|
cmtm6
|
CKLF-like MARVEL transmembrane domain containing 6 |
| chr8_-_32376710 | 3.46 |
ENSDART00000098850
|
lipg
|
lipase, endothelial |
| chr15_-_14706032 | 3.38 |
ENSDART00000164166
|
si:dkey-260j18.2
|
si:dkey-260j18.2 |
| chr23_+_25952724 | 3.37 |
ENSDART00000124963
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr9_+_32838316 | 3.33 |
ENSDART00000172033
|
rabl3
|
RAB, member of RAS oncogene family-like 3 |
| chr15_+_35076414 | 3.29 |
ENSDART00000165210
|
zgc:66024
|
zgc:66024 |
| chr2_+_30198819 | 3.22 |
ENSDART00000004903
|
rdh10b
|
retinol dehydrogenase 10b |
| chr11_-_31012985 | 3.12 |
ENSDART00000162605
|
man2b1
|
mannosidase, alpha, class 2B, member 1 |
| chr1_+_1564451 | 3.08 |
ENSDART00000137184
|
atp1a1a.2
|
ATPase, Na+/K+ transporting, alpha 1a polypeptide, tandem duplicate 2 |
| chr3_-_55252907 | 3.06 |
ENSDART00000108995
|
tex2
|
testis expressed 2 |
| chr8_+_19945528 | 3.05 |
ENSDART00000134124
|
znf692
|
zinc finger protein 692 |
| chr14_+_29932533 | 3.04 |
ENSDART00000017122
|
asah1a
|
N-acylsphingosine amidohydrolase (acid ceramidase) 1a |
| chr15_+_35089305 | 3.02 |
ENSDART00000156515
|
zgc:55621
|
zgc:55621 |
| chr24_-_7928026 | 3.02 |
ENSDART00000145815
|
txndc5
|
thioredoxin domain containing 5 |
| chr2_+_19588034 | 3.00 |
ENSDART00000163875
|
cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr11_-_2343369 | 2.98 |
ENSDART00000167032
ENSDART00000129854 |
tp53rk
|
TP53 regulating kinase |
| chr15_+_21776031 | 2.96 |
ENSDART00000136151
|
NKAPD1
|
zgc:162339 |
| chr5_+_22006830 | 2.90 |
ENSDART00000080919
|
rpl36a
|
ribosomal protein L36A |
| chr21_+_28710341 | 2.90 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr21_+_26954898 | 2.88 |
ENSDART00000114469
|
fkbp2
|
FK506 binding protein 2 |
| chr16_-_17678748 | 2.83 |
ENSDART00000017142
|
m6pr
|
mannose-6-phosphate receptor (cation dependent) |
| chr15_-_18273640 | 2.80 |
ENSDART00000141508
|
btr16
|
bloodthirsty-related gene family, member 16 |
| chr3_+_17901295 | 2.79 |
ENSDART00000035531
|
mettl26
|
methyltransferase like 26 |
| chr2_+_38305689 | 2.79 |
ENSDART00000170672
|
nedd8l
|
neural precursor cell expressed, developmentally down-regulated 8, like |
| chr22_-_11799751 | 2.77 |
ENSDART00000126784
|
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr9_-_705001 | 2.75 |
ENSDART00000147092
|
cflarb
|
CASP8 and FADD-like apoptosis regulator b |
| chr6_+_49724436 | 2.74 |
ENSDART00000154738
|
stx16
|
syntaxin 16 |
| chr6_-_40715613 | 2.71 |
ENSDART00000153702
|
si:ch211-157b11.12
|
si:ch211-157b11.12 |
| chr13_+_29904945 | 2.69 |
ENSDART00000057525
|
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr23_+_25952913 | 2.69 |
ENSDART00000124963
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr11_-_6858626 | 2.68 |
ENSDART00000168372
|
lsm4
|
LSM4 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr21_+_18870471 | 2.67 |
ENSDART00000160185
|
smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr14_-_49798900 | 2.63 |
ENSDART00000169730
|
si:ch211-199b20.3
|
si:ch211-199b20.3 |
| chr2_+_38305772 | 2.61 |
ENSDART00000170672
|
nedd8l
|
neural precursor cell expressed, developmentally down-regulated 8, like |
| chr5_-_57053687 | 2.61 |
ENSDART00000144237
|
gig2p
|
grass carp reovirus (GCRV)-induced gene 2p |
| chr15_+_25554119 | 2.59 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr17_-_24569231 | 2.59 |
ENSDART00000105442
|
aftphb
|
aftiphilin b |
| chr20_-_53123124 | 2.56 |
ENSDART00000162812
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr23_+_25952958 | 2.55 |
ENSDART00000124963
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr18_+_39506453 | 2.53 |
ENSDART00000126978
|
acadl
|
acyl-CoA dehydrogenase, long chain |
| chr2_-_31817448 | 2.52 |
ENSDART00000170880
|
retreg1
|
reticulophagy regulator 1 |
| chr8_+_8605240 | 2.51 |
ENSDART00000075624
|
usp11
|
ubiquitin specific peptidase 11 |
| chr9_-_32994156 | 2.45 |
ENSDART00000133382
|
zgc:112056
|
zgc:112056 |
| chr1_+_11707382 | 2.43 |
ENSDART00000139440
|
zgc:77739
|
zgc:77739 |
| chr6_+_27100350 | 2.42 |
ENSDART00000121558
|
atg4b
|
autophagy related 4B, cysteine peptidase |
| chr3_+_32279283 | 2.40 |
ENSDART00000061426
|
rras
|
related RAS viral (r-ras) oncogene homolog |
| chr3_-_36298676 | 2.40 |
ENSDART00000157950
|
rogdi
|
rogdi homolog (Drosophila) |
| chr18_+_14715573 | 2.37 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr7_+_26274744 | 2.37 |
ENSDART00000135313
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr23_+_30803518 | 2.37 |
ENSDART00000134141
|
asxl1
|
additional sex combs like transcriptional regulator 1 |
| chr13_-_226574 | 2.37 |
|
|
|
| chr19_-_5142209 | 2.35 |
ENSDART00000150980
|
chd4a
|
chromodomain helicase DNA binding protein 4a |
| chr21_+_130546 | 2.32 |
ENSDART00000174654
|
ARSB
|
arylsulfatase B |
| chr2_+_31959127 | 2.31 |
ENSDART00000132113
|
otulinb
|
OTU deubiquitinase with linear linkage specificity b |
| chr4_+_17666992 | 2.31 |
ENSDART00000066999
|
ccdc53
|
coiled-coil domain containing 53 |
| chr5_+_60234640 | 2.31 |
|
|
|
| chr11_+_12983670 | 2.31 |
ENSDART00000166126
|
btf3l4
|
basic transcription factor 3-like 4 |
| chr25_-_3619590 | 2.31 |
ENSDART00000037973
|
morc2
|
MORC family CW-type zinc finger 2 |
| chr5_+_9579430 | 2.30 |
ENSDART00000109236
|
ENSDARG00000075416
|
ENSDARG00000075416 |
| chr2_-_31817409 | 2.29 |
ENSDART00000112763
|
retreg1
|
reticulophagy regulator 1 |
| chr19_+_32178335 | 2.28 |
|
|
|
| chr1_-_11707244 | 2.28 |
ENSDART00000146067
|
sclt1
|
sodium channel and clathrin linker 1 |
| chr18_+_48188699 | 2.27 |
|
|
|
| chr8_+_19945820 | 2.27 |
ENSDART00000134124
|
znf692
|
zinc finger protein 692 |
| chr5_+_51979524 | 2.24 |
ENSDART00000168317
|
apba1a
|
amyloid beta (A4) precursor protein-binding, family A, member 1a |
| chr6_-_55312282 | 2.23 |
ENSDART00000083679
ENSDART00000162117 |
ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr1_+_25411387 | 2.21 |
ENSDART00000127154
|
toporsa
|
topoisomerase I binding, arginine/serine-rich a |
| chr16_-_29755471 | 2.20 |
ENSDART00000133965
|
scnm1
|
sodium channel modifier 1 |
| chr7_-_54752916 | 2.19 |
|
|
|
| chr18_+_6339577 | 2.18 |
ENSDART00000136333
|
wash1
|
WAS protein family homolog 1 |
| chr5_-_25133229 | 2.18 |
ENSDART00000051566
|
zgc:101016
|
zgc:101016 |
| chr4_-_17364090 | 2.18 |
ENSDART00000134467
|
parpbp
|
PARP1 binding protein |
| chr5_-_23525132 | 2.17 |
ENSDART00000051546
|
rps6ka3a
|
ribosomal protein S6 kinase a, polypeptide 3a |
| chr5_-_37648047 | 2.17 |
ENSDART00000139479
|
slc12a9
|
solute carrier family 12, member 9 |
| chr11_-_29740581 | 2.16 |
|
|
|
| chr19_+_21124187 | 2.14 |
ENSDART00000129730
|
rab5aa
|
RAB5A, member RAS oncogene family, a |
| chr7_-_19274727 | 2.14 |
ENSDART00000114203
|
man2b2
|
mannosidase, alpha, class 2B, member 2 |
| chr7_+_26274687 | 2.11 |
ENSDART00000135313
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr19_+_14295978 | 2.10 |
ENSDART00000168260
|
nudc
|
nudC nuclear distribution protein |
| chr19_+_4135556 | 2.10 |
ENSDART00000162461
|
btr26
|
bloodthirsty-related gene family, member 26 |
| chr13_+_11696495 | 2.08 |
ENSDART00000110141
|
sufu
|
suppressor of fused homolog (Drosophila) |
| chr17_+_18217416 | 2.06 |
ENSDART00000156616
|
BX465857.1
|
ENSDARG00000097921 |
| chr13_+_31586024 | 2.06 |
ENSDART00000034745
|
prkcha
|
protein kinase C, eta, a |
| chr3_+_29380031 | 2.06 |
ENSDART00000123619
|
rac2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr21_+_26486084 | 2.05 |
ENSDART00000134617
|
adssl
|
adenylosuccinate synthase, like |
| chr14_-_27040866 | 2.05 |
ENSDART00000173053
|
CU062454.1
|
ENSDARG00000105327 |
| chr17_-_29196678 | 2.04 |
ENSDART00000076481
|
ehd4
|
EH-domain containing 4 |
| chr16_+_21128596 | 2.03 |
ENSDART00000052660
|
skap2
|
src kinase associated phosphoprotein 2 |
| chr25_-_22851190 | 2.03 |
ENSDART00000175228
|
CU462979.1
|
ENSDARG00000107627 |
| chr16_+_43464824 | 2.02 |
ENSDART00000032778
|
rnf144b
|
ring finger protein 144B |
| chr20_-_43853614 | 2.02 |
ENSDART00000100605
|
ttc32
|
tetratricopeptide repeat domain 32 |
| chr12_-_33358480 | 1.99 |
ENSDART00000105562
|
zgc:91940
|
zgc:91940 |
| chr16_-_29517367 | 1.98 |
ENSDART00000148960
|
si:ch211-113g11.6
|
si:ch211-113g11.6 |
| chr7_-_2614300 | 1.98 |
ENSDART00000153548
|
BX323987.1
|
ENSDARG00000097745 |
| chr3_+_32278978 | 1.98 |
ENSDART00000061426
|
rras
|
related RAS viral (r-ras) oncogene homolog |
| chr23_+_24672435 | 1.96 |
ENSDART00000131689
ENSDART00000078796 |
kansl2
|
KAT8 regulatory NSL complex subunit 2 |
| chr14_-_20940726 | 1.95 |
ENSDART00000129743
|
si:ch211-175m2.5
|
si:ch211-175m2.5 |
| chr7_+_23752492 | 1.95 |
ENSDART00000141165
|
tinf2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr7_+_41607231 | 1.95 |
ENSDART00000165789
ENSDART00000115090 |
gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr20_+_27188382 | 1.95 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr10_+_27028355 | 1.94 |
ENSDART00000064111
|
faub
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed b |
| chr20_+_2625716 | 1.93 |
ENSDART00000058775
|
zgc:101562
|
zgc:101562 |
| chr23_-_36349563 | 1.92 |
|
|
|
| chr4_-_20119812 | 1.91 |
ENSDART00000100867
|
fam3c
|
family with sequence similarity 3, member C |
| chr16_-_55210909 | 1.90 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr12_-_24783175 | 1.90 |
|
|
|
| chr7_-_38363533 | 1.90 |
ENSDART00000173861
|
c1qtnf4
|
C1q and TNF related 4 |
| chr20_+_5022501 | 1.89 |
ENSDART00000162678
|
si:dkey-60a16.1
|
si:dkey-60a16.1 |
| chr11_-_2343143 | 1.87 |
ENSDART00000167032
ENSDART00000129854 |
tp53rk
|
TP53 regulating kinase |
| chr5_+_23582676 | 1.87 |
ENSDART00000139520
|
tp53
|
tumor protein p53 |
| chr3_+_36571618 | 1.87 |
ENSDART00000161897
|
abcc1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
| chr11_+_7173167 | 1.86 |
ENSDART00000125619
|
thop1
|
thimet oligopeptidase 1 |
| chr17_+_39842446 | 1.85 |
ENSDART00000149488
|
ubr1
|
ubiquitin protein ligase E3 component n-recognin 1 |
| chr8_+_11649521 | 1.85 |
ENSDART00000167298
|
atp2a2a
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2a |
| chr5_+_1461715 | 1.84 |
ENSDART00000054057
|
ddrgk1
|
DDRGK domain containing 1 |
| chr13_+_29904913 | 1.81 |
ENSDART00000057525
|
dnajb12a
|
DnaJ (Hsp40) homolog, subfamily B, member 12a |
| chr25_+_29245574 | 1.81 |
ENSDART00000073478
|
brd1b
|
bromodomain containing 1b |
| chr6_+_23218068 | 1.79 |
ENSDART00000164960
|
stx8
|
syntaxin 8 |
| chr15_+_12504537 | 1.76 |
ENSDART00000168011
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr10_+_22885419 | 1.75 |
ENSDART00000079437
|
ap4m1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr8_+_15213841 | 1.74 |
ENSDART00000063717
|
ENSDARG00000043402
|
ENSDARG00000043402 |
| chr2_+_35621136 | 1.72 |
ENSDART00000133018
ENSDART00000147278 |
plk3
|
polo-like kinase 3 (Drosophila) |
| chr14_-_16504730 | 1.72 |
ENSDART00000158396
|
tcirg1b
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 subunit A3b |
| chr25_+_16784585 | 1.72 |
ENSDART00000020259
|
zgc:77158
|
zgc:77158 |
| chr18_+_44753053 | 1.72 |
ENSDART00000139690
|
GLB1L2
|
galactosidase beta 1 like 2 |
| chr22_+_2575708 | 1.72 |
|
|
|
| chr19_-_11486168 | 1.71 |
ENSDART00000109889
ENSDART00000172065 |
hsbp1l1
|
heat shock factor binding protein 1 like 1 |
| chr5_-_40524177 | 1.71 |
ENSDART00000083561
|
mtmr12
|
myotubularin related protein 12 |
| chr23_+_43746577 | 1.70 |
ENSDART00000168646
|
slc10a7
|
solute carrier family 10, member 7 |
| KN150262v1_+_4307 | 1.70 |
|
|
|
| chr7_-_13416823 | 1.70 |
ENSDART00000056893
|
pdcd7
|
programmed cell death 7 |
| KN149858v1_+_12702 | 1.70 |
ENSDART00000165916
|
MED8
|
mediator complex subunit 8 |
| chr9_-_32837850 | 1.68 |
ENSDART00000123374
|
dnajb11
|
DnaJ (Hsp40) homolog, subfamily B, member 11 |
| chr20_-_53123015 | 1.67 |
ENSDART00000162812
|
fdft1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr7_-_40307611 | 1.67 |
ENSDART00000173926
ENSDART00000010035 |
dnajb6b
|
DnaJ (Hsp40) homolog, subfamily B, member 6b |
| chr9_+_27909662 | 1.66 |
ENSDART00000112415
|
lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr21_+_28710269 | 1.65 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr3_-_29540088 | 1.65 |
|
|
|
| chr14_+_41039244 | 1.63 |
ENSDART00000111480
|
bcorl1
|
BCL6 corepressor-like 1 |
| chr10_+_19059739 | 1.63 |
ENSDART00000038674
|
tmem230a
|
transmembrane protein 230a |
| chr7_-_69576270 | 1.62 |
|
|
|
| chr9_-_30574226 | 1.62 |
ENSDART00000060134
|
piga
|
phosphatidylinositol glycan anchor biosynthesis, class A |
| chr7_+_24543671 | 1.60 |
ENSDART00000173581
|
si:ch211-226l4.6
|
si:ch211-226l4.6 |
| chr20_-_18482817 | 1.60 |
ENSDART00000159636
|
vipas39
|
VPS33B interacting protein, apical-basolateral polarity regulator, spe-39 homolog |
| chr1_+_11490451 | 1.59 |
ENSDART00000142081
|
stra6l
|
STRA6-like |
| chr22_-_11799717 | 1.59 |
ENSDART00000126784
|
ptpn4b
|
protein tyrosine phosphatase, non-receptor type 4b |
| chr17_-_50140768 | 1.59 |
ENSDART00000058707
|
jdp2a
|
Jun dimerization protein 2a |
| chr2_-_56632969 | 1.59 |
ENSDART00000089158
|
hmha1a
|
histocompatibility (minor) HA-1 a |
| chr17_+_38348229 | 1.58 |
ENSDART00000154346
|
mbip
|
MAP3K12 binding inhibitory protein 1 |
| chr13_+_6908853 | 1.58 |
ENSDART00000109434
|
aifm2
|
apoptosis-inducing factor, mitochondrion-associated, 2 |
| chr14_-_14381338 | 1.57 |
ENSDART00000169970
|
acrc
|
acidic repeat containing |
| chr25_+_35159867 | 1.57 |
|
|
|
| chr7_-_49364447 | 1.56 |
ENSDART00000011456
|
tsg101b
|
tumor susceptibility 101b |
| chr12_+_32316774 | 1.56 |
ENSDART00000153144
|
CT573006.1
|
ENSDARG00000096761 |
| chr8_+_31247312 | 1.56 |
ENSDART00000174921
|
AL929453.2
|
ENSDARG00000108092 |
| chr10_-_35377548 | 1.56 |
ENSDART00000131268
|
rhbdd2
|
rhomboid domain containing 2 |
| chr21_+_28710424 | 1.55 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr3_-_35734943 | 1.55 |
ENSDART00000135957
|
suz12a
|
SUZ12 polycomb repressive complex 2a subunit |
| chr8_+_150970 | 1.55 |
|
|
|
| chr6_+_27100249 | 1.55 |
ENSDART00000121558
|
atg4b
|
autophagy related 4B, cysteine peptidase |
| chr22_-_7688508 | 1.55 |
|
|
|
| chr21_+_130426 | 1.53 |
ENSDART00000174654
|
ARSB
|
arylsulfatase B |
| chr25_-_22851388 | 1.53 |
ENSDART00000175228
|
CU462979.1
|
ENSDARG00000107627 |
| chr1_-_55211204 | 1.53 |
ENSDART00000064194
|
asf1bb
|
anti-silencing function 1Bb histone chaperone |
| chr6_+_94581 | 1.53 |
ENSDART00000125176
|
ap1m2
|
adaptor-related protein complex 1, mu 2 subunit |
| chr5_-_1734159 | 1.53 |
ENSDART00000145781
|
ENSDARG00000094300
|
ENSDARG00000094300 |
| chr8_+_3364453 | 1.52 |
ENSDART00000164426
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr1_-_46423056 | 1.52 |
ENSDART00000053157
|
setd4
|
SET domain containing 4 |
| chr2_+_41909094 | 1.51 |
ENSDART00000179428
|
CABZ01014197.2
|
ENSDARG00000107429 |
| chr8_-_48679386 | 1.51 |
ENSDART00000047134
|
clcn6
|
chloride channel 6 |
| chr23_+_25953162 | 1.51 |
ENSDART00000138731
|
pkig
|
protein kinase (cAMP-dependent, catalytic) inhibitor gamma |
| chr7_-_48532462 | 1.51 |
ENSDART00000015884
|
mfge8a
|
milk fat globule-EGF factor 8 protein a |
| chr18_-_35432838 | 1.51 |
ENSDART00000141703
|
snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr10_+_22885311 | 1.50 |
ENSDART00000079437
|
ap4m1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr22_+_35299551 | 1.50 |
ENSDART00000165353
|
rubcn
|
RUN domain and cysteine-rich domain containing, Beclin 1-interacting protein |
| chr17_-_9794176 | 1.49 |
|
|
|
| chr2_+_38305898 | 1.49 |
ENSDART00000170672
|
nedd8l
|
neural precursor cell expressed, developmentally down-regulated 8, like |
| chr7_+_19348229 | 1.48 |
ENSDART00000007310
ENSDART00000166355 |
zgc:171731
|
zgc:171731 |
| chr8_-_37072857 | 1.48 |
ENSDART00000004041
|
zgc:162200
|
zgc:162200 |
| chr15_+_23592828 | 1.48 |
ENSDART00000152422
|
si:dkey-182i3.8
|
si:dkey-182i3.8 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.4 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 1.6 | 4.8 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 1.4 | 10.1 | GO:2000480 | regulation of cAMP-dependent protein kinase activity(GO:2000479) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.9 | 8.9 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.9 | 4.3 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.8 | 3.3 | GO:0050746 | regulation of lipoprotein metabolic process(GO:0050746) regulation of protein lipidation(GO:1903059) |
| 0.8 | 8.1 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.7 | 5.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.6 | 2.5 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.6 | 2.5 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.6 | 1.9 | GO:1903895 | regulation of endoplasmic reticulum unfolded protein response(GO:1900101) negative regulation of endoplasmic reticulum unfolded protein response(GO:1900102) regulation of IRE1-mediated unfolded protein response(GO:1903894) negative regulation of IRE1-mediated unfolded protein response(GO:1903895) |
| 0.6 | 1.9 | GO:0042908 | xenobiotic transport(GO:0042908) detoxification of inorganic compound(GO:0061687) |
| 0.6 | 4.9 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.6 | 4.2 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.6 | 3.5 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.6 | 1.7 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.6 | 2.8 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) |
| 0.5 | 1.6 | GO:0097435 | fibril organization(GO:0097435) |
| 0.5 | 2.1 | GO:1903533 | regulation of protein targeting(GO:1903533) |
| 0.5 | 1.9 | GO:0010481 | keratinocyte development(GO:0003334) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.5 | 2.3 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.4 | 5.8 | GO:0030011 | maintenance of cell polarity(GO:0030011) maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.4 | 2.7 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.4 | 3.1 | GO:0071939 | vitamin A transport(GO:0071938) vitamin A import(GO:0071939) |
| 0.4 | 2.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.4 | 2.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.4 | 4.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.4 | 2.0 | GO:2000290 | regulation of myotome development(GO:2000290) |
| 0.4 | 1.2 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.4 | 2.4 | GO:1901216 | regulation of transposition, RNA-mediated(GO:0010525) negative regulation of transposition, RNA-mediated(GO:0010526) transposition, RNA-mediated(GO:0032197) positive regulation of neuron apoptotic process(GO:0043525) positive regulation of neuron death(GO:1901216) |
| 0.4 | 6.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.4 | 1.5 | GO:0090140 | regulation of mitochondrial fission(GO:0090140) negative regulation of cell aging(GO:0090344) |
| 0.4 | 6.4 | GO:0035803 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.4 | 1.5 | GO:1903726 | negative regulation of phospholipid metabolic process(GO:1903726) |
| 0.4 | 4.8 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.3 | 3.1 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.3 | 1.4 | GO:0090113 | regulation of COPII vesicle coating(GO:0003400) regulation of ER to Golgi vesicle-mediated transport by GTP hydrolysis(GO:0090113) |
| 0.3 | 1.7 | GO:1903573 | negative regulation of response to endoplasmic reticulum stress(GO:1903573) |
| 0.3 | 1.9 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.3 | 2.8 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.3 | 1.2 | GO:0072401 | signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) signal transduction involved in G2 DNA damage checkpoint(GO:0072425) |
| 0.3 | 6.3 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.3 | 1.7 | GO:0030575 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.3 | 4.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.3 | 1.1 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.3 | 0.8 | GO:0072672 | neutrophil extravasation(GO:0072672) regulation of neutrophil extravasation(GO:2000389) positive regulation of neutrophil extravasation(GO:2000391) |
| 0.3 | 1.6 | GO:0052746 | inositol phosphorylation(GO:0052746) |
| 0.3 | 1.5 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.3 | 1.0 | GO:0030220 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.2 | 1.0 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.2 | 0.9 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.2 | 3.9 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.2 | 1.1 | GO:0032367 | intracellular cholesterol transport(GO:0032367) positive regulation of lipid transport(GO:0032370) |
| 0.2 | 0.9 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.2 | 1.3 | GO:0044528 | regulation of mitochondrial mRNA stability(GO:0044528) |
| 0.2 | 1.1 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.2 | 1.1 | GO:0034367 | macromolecular complex remodeling(GO:0034367) |
| 0.2 | 1.5 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.2 | 0.6 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.2 | 0.9 | GO:0045639 | positive regulation of myeloid cell differentiation(GO:0045639) |
| 0.2 | 3.1 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.2 | 0.5 | GO:0030279 | negative regulation of ossification(GO:0030279) |
| 0.2 | 2.3 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.2 | 0.7 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.2 | 1.0 | GO:0071480 | response to gamma radiation(GO:0010332) cellular response to gamma radiation(GO:0071480) |
| 0.2 | 2.0 | GO:0042113 | B cell activation(GO:0042113) |
| 0.2 | 1.0 | GO:0006750 | glutathione biosynthetic process(GO:0006750) |
| 0.2 | 1.7 | GO:0007035 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.2 | 0.6 | GO:0002931 | response to ischemia(GO:0002931) |
| 0.2 | 0.5 | GO:1903259 | exon-exon junction complex disassembly(GO:1903259) |
| 0.1 | 0.4 | GO:0033238 | regulation of cellular amino acid metabolic process(GO:0006521) regulation of cellular amine metabolic process(GO:0033238) |
| 0.1 | 0.7 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.9 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.7 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 1.5 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.5 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.1 | 2.0 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.1 | 1.5 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 0.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.1 | 0.9 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.1 | 1.0 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.1 | 1.7 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.1 | 0.6 | GO:0099645 | protein localization to postsynaptic specialization membrane(GO:0099633) neurotransmitter receptor localization to postsynaptic specialization membrane(GO:0099645) |
| 0.1 | 0.6 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.8 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.1 | 2.5 | GO:0007338 | single fertilization(GO:0007338) |
| 0.1 | 0.6 | GO:0010971 | regulation of mitotic cell cycle, embryonic(GO:0009794) positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) mitotic cell cycle, embryonic(GO:0045448) positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.1 | 5.7 | GO:0030336 | negative regulation of cell migration(GO:0030336) |
| 0.1 | 0.4 | GO:0018377 | N-terminal protein lipidation(GO:0006498) N-terminal protein myristoylation(GO:0006499) N-terminal peptidyl-glycine N-myristoylation(GO:0018008) protein myristoylation(GO:0018377) |
| 0.1 | 0.5 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 0.9 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.1 | 1.5 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 1.8 | GO:1904356 | regulation of telomere maintenance via telomere lengthening(GO:1904356) |
| 0.1 | 1.4 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.1 | 0.5 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.1 | 2.3 | GO:0051788 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.1 | 1.0 | GO:0006098 | pentose-phosphate shunt(GO:0006098) |
| 0.1 | 0.9 | GO:1902101 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of chromosome segregation(GO:0051984) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.1 | 1.8 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.1 | 3.4 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 1.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.1 | 0.8 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.1 | 1.1 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.1 | 0.6 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 0.5 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.1 | 0.9 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.1 | 1.5 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.1 | 1.1 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.1 | 4.6 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.1 | 1.0 | GO:0007032 | endosome organization(GO:0007032) |
| 0.1 | 0.7 | GO:2000273 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) positive regulation of receptor activity(GO:2000273) |
| 0.1 | 2.7 | GO:1901800 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.1 | 0.3 | GO:0042264 | peptidyl-aspartic acid modification(GO:0018197) peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.1 | 0.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 1.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.1 | 0.5 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 1.0 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 1.0 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 1.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.1 | 4.7 | GO:0006865 | amino acid transport(GO:0006865) |
| 0.1 | 0.6 | GO:0006892 | post-Golgi vesicle-mediated transport(GO:0006892) |
| 0.1 | 0.3 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 1.3 | GO:0043507 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.1 | 2.2 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.1 | 1.6 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.1 | 3.7 | GO:0036503 | ERAD pathway(GO:0036503) |
| 0.1 | 9.8 | GO:0018105 | peptidyl-serine phosphorylation(GO:0018105) |
| 0.1 | 0.6 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.1 | 1.0 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.1 | 1.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.1 | 0.8 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.1 | 1.1 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 1.8 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
| 0.1 | 1.2 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 0.5 | GO:0045841 | mitotic spindle assembly checkpoint(GO:0007094) spindle checkpoint(GO:0031577) negative regulation of mitotic metaphase/anaphase transition(GO:0045841) spindle assembly checkpoint(GO:0071173) mitotic spindle checkpoint(GO:0071174) |
| 0.1 | 0.4 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.1 | 0.8 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 1.5 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.0 | 0.3 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.2 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 1.6 | GO:0051896 | protein kinase B signaling(GO:0043491) regulation of protein kinase B signaling(GO:0051896) |
| 0.0 | 1.4 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.5 | GO:0031063 | regulation of histone deacetylation(GO:0031063) |
| 0.0 | 1.2 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.7 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 1.2 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:0090594 | inflammatory response to wounding(GO:0090594) |
| 0.0 | 1.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 2.8 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.2 | GO:0010889 | regulation of sequestering of triglyceride(GO:0010889) positive regulation of sequestering of triglyceride(GO:0010890) sequestering of triglyceride(GO:0030730) |
| 0.0 | 4.3 | GO:0045087 | innate immune response(GO:0045087) |
| 0.0 | 0.2 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 1.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 1.9 | GO:0008016 | regulation of heart contraction(GO:0008016) |
| 0.0 | 4.6 | GO:0006457 | protein folding(GO:0006457) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 1.6 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.7 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 2.2 | GO:0048232 | male gamete generation(GO:0048232) |
| 0.0 | 2.3 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.9 | GO:0032869 | cellular response to insulin stimulus(GO:0032869) |
| 0.0 | 4.6 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 2.7 | GO:0007626 | locomotory behavior(GO:0007626) |
| 0.0 | 0.7 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 1.2 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) negative regulation of chemotaxis(GO:0050922) negative regulation of axon guidance(GO:1902668) |
| 0.0 | 0.6 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.8 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 3.8 | GO:0043087 | regulation of GTPase activity(GO:0043087) |
| 0.0 | 0.7 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.2 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.5 | GO:0043277 | apoptotic cell clearance(GO:0043277) |
| 0.0 | 0.2 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.7 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.2 | GO:0002574 | thrombocyte differentiation(GO:0002574) |
| 0.0 | 0.8 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.2 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 1.1 | GO:0051321 | meiotic cell cycle(GO:0051321) |
| 0.0 | 0.7 | GO:0006119 | oxidative phosphorylation(GO:0006119) |
| 0.0 | 0.2 | GO:0099518 | vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 0.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.7 | GO:0071559 | transforming growth factor beta receptor signaling pathway(GO:0007179) response to transforming growth factor beta(GO:0071559) cellular response to transforming growth factor beta stimulus(GO:0071560) |
| 0.0 | 0.1 | GO:0032981 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 1.1 | GO:0006413 | translational initiation(GO:0006413) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 8.9 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 1.1 | 7.4 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 1.1 | 3.2 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.8 | 3.9 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.7 | 4.8 | GO:0071546 | pi-body(GO:0071546) |
| 0.5 | 4.9 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.5 | 2.1 | GO:0035301 | Hedgehog signaling complex(GO:0035301) |
| 0.5 | 2.4 | GO:0035517 | PR-DUB complex(GO:0035517) |
| 0.4 | 5.2 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.4 | 1.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.4 | 3.5 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.4 | 1.5 | GO:0002144 | cytosolic tRNA wobble base thiouridylase complex(GO:0002144) |
| 0.3 | 1.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.3 | 0.9 | GO:0097361 | CIA complex(GO:0097361) |
| 0.3 | 4.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.3 | 1.9 | GO:0070187 | telosome(GO:0070187) |
| 0.2 | 8.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.2 | 3.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.2 | 4.3 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.2 | 1.7 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.2 | 1.0 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.2 | 0.6 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.2 | 1.7 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.2 | 0.8 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.2 | 0.5 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.2 | 0.9 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.2 | 1.0 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.1 | 1.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 2.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 2.9 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.1 | 4.4 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 0.9 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 0.7 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.1 | 1.0 | GO:0000974 | Prp19 complex(GO:0000974) U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.1 | 1.5 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 5.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.1 | 0.8 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.1 | 1.2 | GO:0016529 | sarcoplasm(GO:0016528) sarcoplasmic reticulum(GO:0016529) |
| 0.1 | 3.0 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 2.8 | GO:0044309 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.1 | 1.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.1 | 0.7 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.1 | 0.5 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.9 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.7 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 4.8 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.1 | 2.4 | GO:0000777 | condensed chromosome kinetochore(GO:0000777) |
| 0.1 | 11.0 | GO:0005764 | lysosome(GO:0005764) |
| 0.1 | 3.5 | GO:0032592 | integral component of mitochondrial membrane(GO:0032592) |
| 0.0 | 0.6 | GO:0099634 | postsynaptic density membrane(GO:0098839) postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 1.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 3.6 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.8 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 4.4 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 1.2 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.3 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.7 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 2.7 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 2.2 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 4.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 6.0 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.9 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.5 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 1.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 3.7 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
| 0.0 | 0.8 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 5.2 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.1 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.3 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 1.4 | 10.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 1.4 | 4.2 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 1.2 | 4.8 | GO:0034584 | piRNA binding(GO:0034584) |
| 1.1 | 4.3 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 1.0 | 8.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.7 | 4.7 | GO:0022889 | serine transmembrane transporter activity(GO:0022889) |
| 0.6 | 4.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.6 | 2.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.6 | 5.9 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.6 | 3.5 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.5 | 1.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.5 | 1.5 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.5 | 5.6 | GO:0052813 | phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.5 | 1.9 | GO:0035673 | oligopeptide transporter activity(GO:0015198) oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.5 | 1.4 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.5 | 2.8 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.4 | 4.8 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.4 | 1.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.4 | 2.1 | GO:0008506 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.4 | 2.8 | GO:0005537 | mannose binding(GO:0005537) |
| 0.4 | 1.6 | GO:0017064 | fatty acid amide hydrolase activity(GO:0017064) |
| 0.4 | 2.7 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.4 | 6.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.4 | 1.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.3 | 6.9 | GO:0031386 | protein tag(GO:0031386) |
| 0.3 | 2.0 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.3 | 2.5 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.3 | 4.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.3 | 5.9 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.3 | 3.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.3 | 2.1 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.3 | 3.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.3 | 5.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.3 | 6.3 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.3 | 1.6 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.3 | 3.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.3 | 0.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.2 | 1.6 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.2 | 2.7 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 2.9 | GO:0015301 | anion:anion antiporter activity(GO:0015301) |
| 0.2 | 1.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 1.1 | GO:0005163 | nerve growth factor receptor binding(GO:0005163) neurotrophin receptor binding(GO:0005165) |
| 0.2 | 1.9 | GO:0030619 | U1 snRNA binding(GO:0030619) |
| 0.2 | 1.0 | GO:0016429 | tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.2 | 1.7 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.2 | 1.9 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.2 | 1.5 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.2 | 0.5 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.2 | 3.0 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.2 | 1.0 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.2 | 0.7 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.2 | 4.3 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.2 | 0.9 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.1 | 2.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 1.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.1 | 2.3 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 1.5 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 1.0 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.5 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 6.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.1 | 5.6 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.8 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.1 | 0.9 | GO:0043035 | chromatin insulator sequence binding(GO:0043035) |
| 0.1 | 1.9 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 0.9 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.9 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 1.5 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.1 | 0.4 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.4 | GO:0004379 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) |
| 0.1 | 3.0 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.7 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 1.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 0.8 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 1.0 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.1 | 0.1 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.1 | 2.4 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 2.5 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 1.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 8.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.6 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.1 | 1.4 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.1 | 3.7 | GO:0042054 | histone methyltransferase activity(GO:0042054) |
| 0.1 | 7.2 | GO:0101005 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.1 | 2.5 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.1 | 2.7 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.1 | 0.7 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 1.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.1 | 0.5 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 1.0 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.1 | 2.3 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.1 | 0.5 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.1 | 1.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.1 | 4.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.1 | 1.2 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.1 | 0.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 1.9 | GO:0030170 | pyridoxal phosphate binding(GO:0030170) |
| 0.1 | 0.3 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 1.3 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 2.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.7 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 1.3 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.7 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 1.3 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 5.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.9 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 1.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.0 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.7 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 2.7 | GO:1901981 | phosphatidylinositol phosphate binding(GO:1901981) |
| 0.0 | 1.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 2.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.6 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 6.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.9 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 5.9 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 6.4 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 1.3 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 1.0 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 1.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 2.0 | GO:0060090 | binding, bridging(GO:0060090) |
| 0.0 | 0.3 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.8 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.0 | 1.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 1.3 | GO:0015293 | symporter activity(GO:0015293) |
| 0.0 | 0.1 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.0 | 1.2 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 1.3 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.1 | GO:0015141 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) malate transmembrane transporter activity(GO:0015140) succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0016208 | AMP binding(GO:0016208) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.8 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.2 | 6.5 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.2 | 4.7 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.2 | 1.9 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 1.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 2.6 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 2.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.1 | 1.6 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.1 | 0.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 3.3 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 1.3 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.1 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.1 | 0.7 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 2.5 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 1.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 2.5 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.6 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.7 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 2.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.2 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.4 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.8 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.7 | 2.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.5 | 0.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.5 | 3.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.4 | 5.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.3 | 5.7 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.3 | 2.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.3 | 7.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.3 | 4.1 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.2 | 4.7 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.2 | 2.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 2.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.2 | 3.9 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 4.7 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.2 | 1.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.2 | 4.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.2 | 1.6 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.1 | 1.9 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.1 | 2.7 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 1.0 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.3 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.1 | 2.5 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.1 | 0.5 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 0.8 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 3.9 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 0.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 0.6 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 0.9 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.1 | 1.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 1.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 2.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.1 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.1 | 0.2 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.1 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.2 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.1 | 0.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 1.1 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 3.4 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.0 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 2.9 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.9 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 1.1 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.6 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION INITIATION | Genes involved in RNA Polymerase I Transcription Initiation |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.3 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.1 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.2 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.0 | 2.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 1.0 | REACTOME METABOLISM OF CARBOHYDRATES | Genes involved in Metabolism of carbohydrates |