Project
DANIO-CODE
Navigation
Downloads
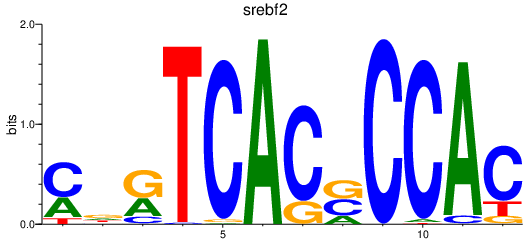
Results for srebf2
Z-value: 1.28
Transcription factors associated with srebf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
srebf2
|
ENSDARG00000063438 | sterol regulatory element binding transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| srebf2 | dr10_dc_chr3_+_1338347_1338547 | 0.75 | 8.2e-04 | Click! |
Activity profile of srebf2 motif
Sorted Z-values of srebf2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of srebf2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_24747855 | 3.82 |
ENSDART00000111686
|
fhdc1
|
FH2 domain containing 1 |
| chr16_-_29229687 | 3.66 |
ENSDART00000132589
|
mef2d
|
myocyte enhancer factor 2d |
| chr9_-_7560915 | 3.42 |
ENSDART00000081553
|
desma
|
desmin a |
| chr7_+_25649559 | 3.24 |
ENSDART00000026295
|
arrb2b
|
arrestin, beta 2b |
| chr21_+_28408329 | 3.21 |
ENSDART00000077871
|
pygma
|
phosphorylase, glycogen, muscle A |
| chr7_+_39130411 | 3.09 |
ENSDART00000144750
|
tnni2b.1
|
troponin I type 2b (skeletal, fast), tandem duplicate 1 |
| chr9_+_907908 | 3.08 |
ENSDART00000144114
|
dbi
|
diazepam binding inhibitor (GABA receptor modulator, acyl-CoA binding protein) |
| chr13_-_33268447 | 2.99 |
ENSDART00000103628
|
btbd6a
|
BTB (POZ) domain containing 6a |
| chr10_+_4874841 | 2.80 |
ENSDART00000165942
|
palm2
|
paralemmin 2 |
| chr22_-_13139894 | 2.75 |
ENSDART00000105762
|
ahr2
|
aryl hydrocarbon receptor 2 |
| chr21_-_18969970 | 2.74 |
ENSDART00000080269
|
pgam2
|
phosphoglycerate mutase 2 (muscle) |
| chr21_-_1716495 | 2.55 |
ENSDART00000151049
|
onecut2
|
one cut homeobox 2 |
| chr9_+_19087329 | 2.40 |
ENSDART00000110457
|
crfb1
|
cytokine receptor family member b1 |
| chr3_-_60866373 | 2.32 |
ENSDART00000112043
|
cacng4b
|
calcium channel, voltage-dependent, gamma subunit 4b |
| chr13_-_2259405 | 2.29 |
ENSDART00000166456
|
elovl5
|
ELOVL fatty acid elongase 5 |
| chr6_-_34974860 | 2.27 |
ENSDART00000113097
|
hsd17b7
|
hydroxysteroid (17-beta) dehydrogenase 7 |
| chr2_+_19546837 | 2.26 |
ENSDART00000163137
|
elovl1a
|
ELOVL fatty acid elongase 1a |
| chr12_-_22888090 | 1.89 |
ENSDART00000111801
ENSDART00000149474 |
mkxa
|
mohawk homeobox a |
| chr23_-_20332076 | 1.87 |
ENSDART00000147326
|
lamb2
|
laminin, beta 2 (laminin S) |
| chr6_-_15630912 | 1.80 |
ENSDART00000063665
|
ackr3b
|
atypical chemokine receptor 3b |
| KN150108v1_+_18276 | 1.79 |
|
|
|
| chr7_-_29811734 | 1.78 |
ENSDART00000075600
|
tspan3b
|
tetraspanin 3b |
| chr16_+_1302842 | 1.76 |
|
|
|
| chr15_+_30276801 | 1.73 |
ENSDART00000047248
ENSDART00000123937 |
nlk2
|
nemo-like kinase, type 2 |
| chr11_-_1792616 | 1.65 |
|
|
|
| chr17_-_6580876 | 1.64 |
ENSDART00000157125
|
vsnl1b
|
visinin-like 1b |
| chr23_-_7740845 | 1.53 |
ENSDART00000172451
|
plagl2
|
pleiomorphic adenoma gene-like 2 |
| chr3_-_35671276 | 1.51 |
ENSDART00000150911
|
caskin1
|
CASK interacting protein 1 |
| chr4_-_44105748 | 1.47 |
ENSDART00000150621
|
znf1137
|
zinc finger protein 1137 |
| chr16_-_204751 | 1.30 |
|
|
|
| chr4_+_632652 | 1.27 |
ENSDART00000171138
|
xpot
|
exportin, tRNA (nuclear export receptor for tRNAs) |
| chr20_-_27412546 | 1.25 |
ENSDART00000148361
|
asb2a.1
|
ankyrin repeat and SOCS box containing 2a, tandem duplicate 1 |
| chr9_-_30553051 | 1.24 |
ENSDART00000147030
|
sytl5
|
synaptotagmin-like 5 |
| chr13_+_21695882 | 1.22 |
ENSDART00000165150
|
zswim8
|
zinc finger, SWIM-type containing 8 |
| chr19_-_46058123 | 1.14 |
|
|
|
| chr1_-_20132900 | 1.06 |
ENSDART00000054472
|
tll1
|
tolloid-like 1 |
| chr24_-_28150352 | 1.05 |
ENSDART00000145290
|
bcl2a
|
B-cell CLL/lymphoma 2a |
| chr3_+_23590779 | 1.04 |
ENSDART00000012470
|
hoxb4a
|
homeobox B4a |
| chr25_-_4296732 | 1.01 |
|
|
|
| chr7_+_44106327 | 0.99 |
ENSDART00000108766
ENSDART00000111441 |
cdh5
|
cadherin 5 |
| chr5_+_36431907 | 0.99 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr5_+_36431824 | 0.99 |
ENSDART00000045036
|
nfkbib
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr6_+_51676410 | 0.97 |
ENSDART00000151565
|
fam65c
|
family with sequence similarity 65, member C |
| chr24_-_3445036 | 0.95 |
ENSDART00000143723
|
idi1
|
isopentenyl-diphosphate delta isomerase 1 |
| chr1_-_9256864 | 0.92 |
ENSDART00000041849
|
tmem8a
|
transmembrane protein 8A |
| chr8_+_20589483 | 0.90 |
ENSDART00000088668
|
nfic
|
nuclear factor I/C |
| chr3_-_47356049 | 0.86 |
ENSDART00000155324
|
pdpk1a
|
3-phosphoinositide dependent protein kinase 1a |
| chr23_-_18131105 | 0.85 |
ENSDART00000173102
|
zgc:92287
|
zgc:92287 |
| chr24_-_3445303 | 0.85 |
ENSDART00000143723
|
idi1
|
isopentenyl-diphosphate delta isomerase 1 |
| chr8_-_26942548 | 0.81 |
ENSDART00000099214
|
slc16a1b
|
solute carrier family 16 (monocarboxylate transporter), member 1b |
| chr16_-_29229755 | 0.76 |
ENSDART00000132589
|
mef2d
|
myocyte enhancer factor 2d |
| chr1_-_29885008 | 0.73 |
ENSDART00000018827
|
dachc
|
dachshund c |
| chr12_-_34358536 | 0.73 |
ENSDART00000111717
|
fscn2b
|
fascin actin-bundling protein 2b, retinal |
| chr5_-_14990023 | 0.70 |
|
|
|
| chr12_+_19262722 | 0.67 |
ENSDART00000078266
|
rsl1d1
|
ribosomal L1 domain containing 1 |
| chr10_+_4874800 | 0.64 |
ENSDART00000165942
|
palm2
|
paralemmin 2 |
| chr5_+_35913418 | 0.55 |
ENSDART00000013590
|
wnt11
|
wingless-type MMTV integration site family, member 11 |
| chr1_+_13342678 | 0.43 |
|
|
|
| chr2_-_23330863 | 0.42 |
|
|
|
| chr10_+_22066137 | 0.40 |
ENSDART00000143461
|
npm1a
|
nucleophosmin 1a (nucleolar phosphoprotein B23, numatrin) |
| chr3_+_1338347 | 0.37 |
ENSDART00000092690
ENSDART00000165395 |
srebf2
|
sterol regulatory element binding transcription factor 2 |
| chr9_-_29687072 | 0.37 |
ENSDART00000060246
|
dnajc3a
|
DnaJ (Hsp40) homolog, subfamily C, member 3a |
| chr19_-_10277398 | 0.31 |
ENSDART00000091707
|
dbpa
|
D site albumin promoter binding protein a |
| chr25_-_17271146 | 0.30 |
ENSDART00000064591
|
cyp2x6
|
cytochrome P450, family 2, subfamily X, polypeptide 6 |
| chr20_+_38955797 | 0.28 |
|
|
|
| chr20_-_7186902 | 0.27 |
ENSDART00000012247
|
dhcr24
|
24-dehydrocholesterol reductase |
| chr2_+_34155114 | 0.25 |
ENSDART00000137170
|
dars2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chr1_+_8873083 | 0.17 |
ENSDART00000111131
|
elfn1b
|
extracellular leucine-rich repeat and fibronectin type III domain containing 1b |
| chr5_-_13844114 | 0.17 |
|
|
|
| chr11_-_3266080 | 0.12 |
ENSDART00000154314
|
prph
|
peripherin |
| chr23_-_18131038 | 0.11 |
ENSDART00000173385
|
zgc:92287
|
zgc:92287 |
| chr5_+_35913494 | 0.02 |
ENSDART00000013590
|
wnt11
|
wingless-type MMTV integration site family, member 11 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.2 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) regulation of protein binding(GO:0043393) |
| 0.8 | 2.3 | GO:0098943 | neurotransmitter receptor transport, postsynaptic endosome to lysosome(GO:0098943) |
| 0.7 | 2.7 | GO:0042698 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) |
| 0.6 | 3.4 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.5 | 1.8 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) |
| 0.5 | 1.8 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.4 | 4.4 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.3 | 1.0 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.3 | 3.2 | GO:0044247 | polysaccharide catabolic process(GO:0000272) glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.3 | 1.3 | GO:0051031 | tRNA transport(GO:0051031) |
| 0.2 | 2.3 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.2 | 0.8 | GO:0035124 | embryonic caudal fin morphogenesis(GO:0035124) |
| 0.2 | 1.9 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 2.3 | GO:1902653 | cholesterol biosynthetic process(GO:0006695) secondary alcohol biosynthetic process(GO:1902653) |
| 0.1 | 3.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 1.9 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.1 | 2.3 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.4 | GO:0006407 | rRNA export from nucleus(GO:0006407) rRNA transport(GO:0051029) |
| 0.1 | 2.7 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 1.1 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.1 | 2.4 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 0.4 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.1 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 1.7 | GO:0030901 | midbrain development(GO:0030901) |
| 0.0 | 0.7 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 1.2 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.0 | 0.8 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.7 | GO:0051017 | actin filament bundle assembly(GO:0051017) |
| 0.0 | 0.6 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 3.0 | GO:0050767 | regulation of neurogenesis(GO:0050767) |
| 0.0 | 0.2 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.2 | 2.3 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.2 | 1.9 | GO:0043256 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 2.7 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 3.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.3 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 3.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 4.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 1.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.8 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.7 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 1.1 | GO:0031965 | nuclear membrane(GO:0031965) |
| 0.0 | 0.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.2 | GO:0004645 | phosphorylase activity(GO:0004645) glycogen phosphorylase activity(GO:0008184) |
| 0.5 | 2.7 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) |
| 0.3 | 1.8 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.3 | 1.8 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.2 | 4.6 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 3.1 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 0.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 2.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 1.7 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 2.4 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.1 | 1.9 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 2.7 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.0 | 1.3 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 2.3 | GO:0016616 | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor(GO:0016616) |
| 0.0 | 0.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 1.0 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 5.9 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 0.7 | GO:0030674 | protein binding, bridging(GO:0030674) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 4.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.1 | 2.0 | ST GA13 PATHWAY | G alpha 13 Pathway |
| 0.0 | 0.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.3 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.2 | 4.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.2 | 2.7 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.2 | 2.0 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.1 | 4.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.6 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |