Project
DANIO-CODE
Navigation
Downloads
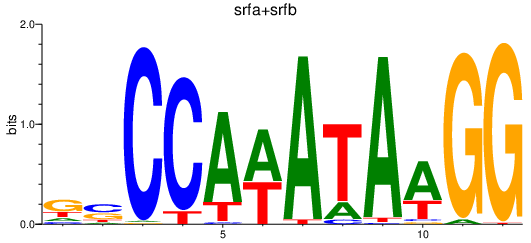
Results for srfa+srfb
Z-value: 3.12
Transcription factors associated with srfa+srfb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
srfa
|
ENSDARG00000053918 | serum response factor a |
|
srfb
|
ENSDARG00000102867 | serum response factor b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| srfa | dr10_dc_chr22_+_35113233_35113370 | 0.84 | 5.4e-05 | Click! |
Activity profile of srfa+srfb motif
Sorted Z-values of srfa+srfb motif
Network of associatons between targets according to the STRING database.
First level regulatory network of srfa+srfb
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_53286155 | 16.95 |
ENSDART00000139625
|
acta1a
|
actin, alpha 1a, skeletal muscle |
| chr7_+_29680967 | 15.65 |
ENSDART00000173540
|
tpma
|
alpha-tropomyosin |
| chr20_-_9992898 | 14.87 |
ENSDART00000080664
|
ACTC1
|
zgc:86709 |
| chr5_-_31708933 | 14.37 |
ENSDART00000123003
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr5_-_31674204 | 14.24 |
ENSDART00000122889
|
myhz1.3
|
myosin, heavy polypeptide 1.3, skeletal muscle |
| chr14_-_21039158 | 14.01 |
ENSDART00000054460
|
egr1
|
early growth response 1 |
| chr13_+_24148599 | 13.50 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr5_+_36487425 | 12.42 |
ENSDART00000049900
|
tagln2
|
transgelin 2 |
| chr20_-_29517770 | 12.28 |
ENSDART00000147464
|
ryr3
|
ryanodine receptor 3 |
| chr11_+_6118409 | 10.66 |
ENSDART00000027666
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr8_-_11191783 | 10.32 |
ENSDART00000131171
|
unc45b
|
unc-45 myosin chaperone B |
| chr12_-_25973094 | 9.10 |
ENSDART00000171206
ENSDART00000171212 ENSDART00000170265 |
ldb3b
|
LIM domain binding 3b |
| chr5_-_31692803 | 8.93 |
ENSDART00000131983
|
myhz1.2
|
myosin, heavy polypeptide 1.2, skeletal muscle |
| chr2_+_30932612 | 8.05 |
ENSDART00000132450
ENSDART00000137012 |
myom1a
|
myomesin 1a (skelemin) |
| chr12_+_42281025 | 7.95 |
ENSDART00000167324
|
ebf3a
|
early B-cell factor 3a |
| chr22_-_36915567 | 7.94 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr10_+_6316944 | 7.72 |
ENSDART00000162428
|
tpm2
|
tropomyosin 2 (beta) |
| chr3_+_23577103 | 7.64 |
ENSDART00000025449
|
hoxb5a
|
homeobox B5a |
| chr5_+_49093250 | 7.34 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr2_-_24898678 | 7.26 |
ENSDART00000145145
|
cnn2
|
calponin 2 |
| chr6_-_39315024 | 7.17 |
ENSDART00000012644
|
krt4
|
keratin 4 |
| chr19_-_6384776 | 7.04 |
|
|
|
| chr13_-_3022033 | 6.80 |
ENSDART00000050934
|
pkdcca
|
protein kinase domain containing, cytoplasmic a |
| chr5_+_49093134 | 6.79 |
ENSDART00000133384
|
nr2f1a
|
nuclear receptor subfamily 2, group F, member 1a |
| chr1_+_51925783 | 6.69 |
ENSDART00000167514
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr1_+_1657493 | 6.57 |
ENSDART00000103850
|
atp1a1a.3
|
ATPase, Na+/K+ transporting, alpha 1a polypeptide, tandem duplicate 3 |
| chr21_+_25729090 | 6.56 |
ENSDART00000021664
|
cldnb
|
claudin b |
| chr20_-_42805703 | 6.36 |
ENSDART00000045816
|
plg
|
plasminogen |
| chr18_-_23889256 | 6.06 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr20_+_14979154 | 5.91 |
ENSDART00000118157
|
dre-mir-199-1
|
dre-mir-199-1 |
| chr7_-_71566426 | 5.84 |
ENSDART00000166724
|
myom1b
|
myomesin 1b |
| chr14_+_45895103 | 5.81 |
|
|
|
| chr16_+_53178713 | 5.76 |
ENSDART00000174634
|
CABZ01062546.1
|
ENSDARG00000108858 |
| chr9_-_47833777 | 5.76 |
|
|
|
| chr8_+_20125687 | 5.20 |
ENSDART00000124809
|
acsbg2
|
acyl-CoA synthetase bubblegum family member 2 |
| chr1_-_50147413 | 5.17 |
ENSDART00000080389
|
fam13a
|
family with sequence similarity 13, member A |
| chr14_+_34174018 | 5.16 |
ENSDART00000024440
|
foxi3b
|
forkhead box I3b |
| chr12_+_19240407 | 5.04 |
ENSDART00000041711
|
gspt1
|
G1 to S phase transition 1 |
| chr11_+_6118322 | 4.86 |
ENSDART00000165031
|
nr2f6b
|
nuclear receptor subfamily 2, group F, member 6b |
| chr22_-_15576473 | 4.82 |
ENSDART00000036075
|
tpm4a
|
tropomyosin 4a |
| chr2_-_55879949 | 4.80 |
ENSDART00000140309
|
cplx4c
|
complexin 4c |
| chr23_+_42920817 | 4.78 |
ENSDART00000055577
|
myl9a
|
myosin, light chain 9a, regulatory |
| chr9_-_47833718 | 4.77 |
|
|
|
| chr7_-_28278286 | 4.71 |
ENSDART00000113313
|
st5
|
suppression of tumorigenicity 5 |
| chr20_+_40247918 | 4.70 |
ENSDART00000121818
|
trdn
|
triadin |
| chr20_+_35377079 | 4.68 |
ENSDART00000168216
|
fam49a
|
family with sequence similarity 49, member A |
| chr13_+_29108288 | 4.63 |
ENSDART00000026000
|
myofl
|
myoferlin like |
| chr21_+_6858073 | 4.53 |
ENSDART00000139493
|
olfm1b
|
olfactomedin 1b |
| chr23_-_7283514 | 4.38 |
ENSDART00000156369
|
CR589876.1
|
ENSDARG00000096997 |
| chr6_-_18733424 | 4.37 |
ENSDART00000151578
|
tns1a
|
tensin 1a |
| chr18_-_23888988 | 4.25 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr13_-_3022193 | 4.19 |
ENSDART00000050934
|
pkdcca
|
protein kinase domain containing, cytoplasmic a |
| chr5_+_37861949 | 4.18 |
ENSDART00000144425
|
gltpd2
|
glycolipid transfer protein domain containing 2 |
| chr18_+_35433242 | 4.17 |
ENSDART00000057733
|
zgc:153119
|
zgc:153119 |
| chr24_+_31340115 | 4.16 |
ENSDART00000165993
|
f3a
|
coagulation factor IIIa |
| chr18_-_23889025 | 4.13 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr2_-_24614261 | 4.11 |
ENSDART00000099532
|
vmhc
|
ventricular myosin heavy chain |
| chr21_-_17566461 | 3.94 |
ENSDART00000044078
ENSDART00000168495 ENSDART00000020048 |
gsna
|
gelsolin a |
| chr13_-_3022131 | 3.82 |
ENSDART00000050934
|
pkdcca
|
protein kinase domain containing, cytoplasmic a |
| chr22_-_36915720 | 3.79 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr25_+_32707410 | 3.78 |
ENSDART00000125892
ENSDART00000121680 |
ENSDARG00000087402
|
ENSDARG00000087402 |
| chr7_-_52848084 | 3.77 |
ENSDART00000172179
ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr23_-_10241885 | 3.77 |
ENSDART00000081223
ENSDART00000144280 |
krt5
|
keratin 5 |
| chr3_-_35472632 | 3.77 |
ENSDART00000055269
|
gng13b
|
guanine nucleotide binding protein (G protein), gamma 13b |
| chr13_+_24148474 | 3.76 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr24_+_40292617 | 3.64 |
|
|
|
| chr23_+_36019989 | 3.63 |
ENSDART00000103134
ENSDART00000139319 |
hoxc5a
|
homeobox C5a |
| chr16_+_28449134 | 3.61 |
ENSDART00000059044
|
itga8
|
integrin, alpha 8 |
| chr12_+_27038210 | 3.58 |
ENSDART00000076161
|
hoxb5b
|
homeobox B5b |
| chr20_+_20287015 | 3.49 |
ENSDART00000002507
|
rhoj
|
ras homolog family member J |
| chr8_+_50738477 | 3.36 |
ENSDART00000127062
|
egr3
|
early growth response 3 |
| chr18_-_23889169 | 3.35 |
ENSDART00000059976
|
nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr19_+_32679320 | 3.31 |
|
|
|
| chr1_-_5796394 | 3.25 |
ENSDART00000109356
|
klf7a
|
Kruppel-like factor 7a |
| chr19_+_32679258 | 3.07 |
|
|
|
| chr7_+_5372830 | 3.03 |
ENSDART00000135271
|
MYADM
|
myeloid associated differentiation marker |
| chr22_-_29387056 | 3.00 |
ENSDART00000121599
|
pdgfba
|
platelet-derived growth factor beta polypeptide a |
| chr10_-_42157835 | 2.97 |
ENSDART00000141500
ENSDART00000156626 |
BX005309.1
|
ENSDARG00000093535 |
| chr16_+_12740993 | 2.95 |
ENSDART00000128497
|
nat14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr22_+_35113233 | 2.94 |
ENSDART00000123066
|
srfa
|
serum response factor a |
| chr12_+_27038298 | 2.93 |
ENSDART00000076161
|
hoxb5b
|
homeobox B5b |
| chr17_-_20098393 | 2.90 |
ENSDART00000165606
|
ryr2b
|
ryanodine receptor 2b (cardiac) |
| chr2_+_24718602 | 2.89 |
ENSDART00000022379
|
nr2f6a
|
nuclear receptor subfamily 2, group F, member 6a |
| chr23_+_36019758 | 2.84 |
ENSDART00000103134
ENSDART00000139319 |
hoxc5a
|
homeobox C5a |
| chr9_-_39190534 | 2.76 |
|
|
|
| chr5_+_69401770 | 2.74 |
|
|
|
| chr10_+_44509370 | 2.68 |
|
|
|
| chr10_-_44509732 | 2.66 |
|
|
|
| chr5_+_35198499 | 2.53 |
|
|
|
| chr14_-_25011874 | 2.49 |
ENSDART00000159569
|
htr4
|
5-hydroxytryptamine receptor 4 |
| chr17_-_42778156 | 2.49 |
ENSDART00000140549
|
nkx2.2a
|
NK2 homeobox 2a |
| chr5_-_54773058 | 2.48 |
ENSDART00000145791
|
prune2
|
prune homolog 2 (Drosophila) |
| KN150015v1_+_1935 | 2.40 |
ENSDART00000169907
ENSDART00000160155 |
PDCL3
|
phosducin like 3 |
| chr20_+_53772078 | 2.28 |
ENSDART00000126983
|
myh6
|
myosin, heavy chain 6, cardiac muscle, alpha |
| chr19_-_34328673 | 2.27 |
ENSDART00000164563
|
elmo1
|
engulfment and cell motility 1 (ced-12 homolog, C. elegans) |
| chr7_+_29681196 | 2.21 |
ENSDART00000039657
|
tpma
|
alpha-tropomyosin |
| chr20_+_14978952 | 2.13 |
ENSDART00000118157
|
dre-mir-199-1
|
dre-mir-199-1 |
| chr7_+_29680933 | 2.12 |
ENSDART00000173540
|
tpma
|
alpha-tropomyosin |
| chr22_-_18520027 | 2.12 |
ENSDART00000132156
|
cirbpb
|
cold inducible RNA binding protein b |
| chr10_-_29930133 | 2.10 |
ENSDART00000055913
|
hist2h2l
|
histone 2, H2, like |
| chr20_+_6600246 | 2.10 |
ENSDART00000136567
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr2_-_55879979 | 2.10 |
ENSDART00000150179
|
cplx4c
|
complexin 4c |
| chr3_-_22098046 | 2.00 |
ENSDART00000017750
|
myl4
|
myosin, light chain 4, alkali; atrial, embryonic |
| chr17_-_20959288 | 2.00 |
ENSDART00000006676
|
phyhipla
|
phytanoyl-CoA 2-hydroxylase interacting protein-like a |
| chr12_+_42281154 | 1.99 |
ENSDART00000167324
|
ebf3a
|
early B-cell factor 3a |
| chr20_-_2936289 | 1.98 |
ENSDART00000104667
|
cdk19
|
cyclin-dependent kinase 19 |
| chr10_-_1690605 | 1.93 |
ENSDART00000137620
|
si:ch73-46j18.5
|
si:ch73-46j18.5 |
| chr5_-_70991958 | 1.88 |
ENSDART00000011955
ENSDART00000160380 |
gpsm1b
|
G protein signaling modulator 1b |
| chr21_+_6858020 | 1.88 |
ENSDART00000139493
|
olfm1b
|
olfactomedin 1b |
| chr20_+_6600184 | 1.62 |
ENSDART00000136567
|
tns3.2
|
tensin 3, tandem duplicate 2 |
| chr4_-_16655742 | 1.55 |
ENSDART00000042307
|
fam60a
|
family with sequence similarity 60, member A |
| chr3_-_40387858 | 1.40 |
ENSDART00000055194
|
actb2
|
actin, beta 2 |
| chr11_-_11722452 | 1.39 |
ENSDART00000161293
|
CR848705.2
|
ENSDARG00000104692 |
| chr22_-_15576405 | 1.39 |
ENSDART00000009054
|
tpm4a
|
tropomyosin 4a |
| chr12_+_16978590 | 1.38 |
ENSDART00000177923
|
acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr17_-_8498935 | 1.37 |
ENSDART00000148990
|
ctbp2a
|
C-terminal binding protein 2a |
| chr20_-_48255596 | 1.33 |
ENSDART00000031167
|
tfap2d
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr14_+_45895037 | 1.32 |
|
|
|
| chr3_+_58096720 | 1.32 |
ENSDART00000010395
ENSDART00000159755 ENSDART00000171149 |
uqcrc2a
|
ubiquinol-cytochrome c reductase core protein IIa |
| chr22_-_36915689 | 1.29 |
ENSDART00000029588
|
kng1
|
kininogen 1 |
| chr20_+_54544931 | 1.26 |
|
|
|
| chr20_+_40248154 | 1.22 |
ENSDART00000121818
|
trdn
|
triadin |
| chr21_-_2597493 | 1.22 |
|
|
|
| chr11_+_16018035 | 1.19 |
ENSDART00000081054
|
tada3l
|
transcriptional adaptor 3 (NGG1 homolog, yeast)-like |
| chr7_+_71572830 | 1.14 |
ENSDART00000171996
|
myl12.1
|
myosin, light chain 12, genome duplicate 1 |
| chr20_+_35377155 | 1.09 |
ENSDART00000147320
ENSDART00000131974 |
fam49a
|
family with sequence similarity 49, member A |
| chr3_-_51038166 | 1.06 |
|
|
|
| chr9_+_6609374 | 1.04 |
ENSDART00000122279
|
fhl2a
|
four and a half LIM domains 2a |
| chr16_-_30463239 | 1.03 |
ENSDART00000003752
|
cct3
|
chaperonin containing TCP1, subunit 3 (gamma) |
| chr12_-_8633437 | 1.02 |
|
|
|
| chr2_-_49149839 | 0.98 |
|
|
|
| chr5_-_31708973 | 0.96 |
ENSDART00000123003
|
myhz1.1
|
myosin, heavy polypeptide 1.1, skeletal muscle |
| chr15_-_39783178 | 0.89 |
|
|
|
| chr1_-_7968497 | 0.84 |
ENSDART00000141407
ENSDART00000148067 |
actb1
|
actin, beta 1 |
| chr14_+_4169371 | 0.69 |
ENSDART00000136665
|
gnpda2
|
glucosamine-6-phosphate deaminase 2 |
| chr20_+_37963275 | 0.64 |
ENSDART00000153190
|
vash2
|
vasohibin 2 |
| chr19_+_7630847 | 0.60 |
ENSDART00000151758
|
pbxip1a
|
pre-B-cell leukemia homeobox interacting protein 1a |
| chr7_-_28278230 | 0.56 |
ENSDART00000054368
|
st5
|
suppression of tumorigenicity 5 |
| chr22_+_33408025 | 0.52 |
ENSDART00000101580
|
nicn1
|
nicolin 1 |
| chr12_-_26759201 | 0.52 |
|
|
|
| chr23_+_36020128 | 0.52 |
ENSDART00000103134
ENSDART00000139319 |
hoxc5a
|
homeobox C5a |
| chr19_+_46567459 | 0.51 |
ENSDART00000164055
|
cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr23_+_36019942 | 0.51 |
ENSDART00000103134
ENSDART00000139319 |
hoxc5a
|
homeobox C5a |
| chr2_+_56045488 | 0.49 |
ENSDART00000021011
|
upf1
|
upf1 regulator of nonsense transcripts homolog (yeast) |
| chr14_+_300285 | 0.43 |
ENSDART00000159949
|
wdr1
|
WD repeat domain 1 |
| chr13_+_24148390 | 0.28 |
ENSDART00000058629
|
acta1b
|
actin, alpha 1b, skeletal muscle |
| chr10_+_8143021 | 0.22 |
ENSDART00000138875
|
pstpip2
|
proline-serine-threonine phosphatase interacting protein 2 |
| chr22_-_18520332 | 0.21 |
|
|
|
| chr1_-_53081860 | 0.18 |
ENSDART00000026409
|
cct4
|
chaperonin containing TCP1, subunit 4 (delta) |
| chr19_+_44120313 | 0.16 |
ENSDART00000006344
|
cap1
|
CAP, adenylate cyclase-associated protein 1 (yeast) |
| chr3_+_51182773 | 0.16 |
|
|
|
| chr23_-_16765933 | 0.14 |
ENSDART00000046784
|
fkbp1ab
|
FK506 binding protein 1Ab |
| chr1_-_7967515 | 0.10 |
ENSDART00000133319
|
actb1
|
actin, beta 1 |
| chr10_-_1690504 | 0.06 |
ENSDART00000137620
|
si:ch73-46j18.5
|
si:ch73-46j18.5 |
| chr16_-_2880109 | 0.03 |
ENSDART00000149463
|
hace1
|
HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 17.8 | GO:0060845 | lymphatic endothelial cell fate commitment(GO:0060838) venous endothelial cell differentiation(GO:0060843) venous endothelial cell fate commitment(GO:0060845) |
| 2.8 | 19.4 | GO:0030195 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 2.6 | 18.3 | GO:0072132 | mesenchyme morphogenesis(GO:0072132) |
| 1.7 | 5.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 1.5 | 16.4 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) |
| 1.1 | 6.7 | GO:0032374 | regulation of sterol transport(GO:0032371) regulation of cholesterol transport(GO:0032374) |
| 1.0 | 5.0 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.8 | 2.5 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.7 | 15.3 | GO:0001757 | somite specification(GO:0001757) |
| 0.6 | 2.5 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.6 | 7.3 | GO:0043535 | regulation of blood vessel endothelial cell migration(GO:0043535) |
| 0.6 | 15.2 | GO:0051209 | release of sequestered calcium ion into cytosol(GO:0051209) |
| 0.5 | 6.5 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.5 | 5.2 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.5 | 3.8 | GO:0007606 | sensory perception of chemical stimulus(GO:0007606) |
| 0.4 | 2.5 | GO:0006797 | polyphosphate metabolic process(GO:0006797) polyphosphate catabolic process(GO:0006798) |
| 0.4 | 5.8 | GO:0045580 | regulation of T cell differentiation(GO:0045580) |
| 0.4 | 43.0 | GO:0006936 | muscle contraction(GO:0006936) |
| 0.4 | 6.6 | GO:0030007 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.4 | 3.0 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.4 | 14.0 | GO:0032922 | circadian regulation of gene expression(GO:0032922) |
| 0.3 | 1.4 | GO:0070073 | clustering of voltage-gated calcium channels(GO:0070073) |
| 0.3 | 5.9 | GO:0002028 | regulation of sodium ion transport(GO:0002028) |
| 0.3 | 10.5 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.3 | 6.9 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.2 | 2.5 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 4.6 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.1 | 8.1 | GO:0014904 | myotube cell development(GO:0014904) skeletal muscle fiber development(GO:0048741) |
| 0.1 | 3.9 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.1 | 0.7 | GO:0006043 | glucosamine metabolic process(GO:0006041) glucosamine catabolic process(GO:0006043) |
| 0.1 | 0.7 | GO:0030817 | regulation of cAMP metabolic process(GO:0030814) regulation of cAMP biosynthetic process(GO:0030817) regulation of adenylate cyclase activity(GO:0045761) |
| 0.1 | 7.6 | GO:0001708 | cell fate specification(GO:0001708) |
| 0.1 | 17.1 | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter(GO:0000122) |
| 0.1 | 2.3 | GO:0045766 | positive regulation of angiogenesis(GO:0045766) |
| 0.1 | 3.6 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 3.7 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.0 | 1.0 | GO:2000027 | regulation of organ morphogenesis(GO:2000027) |
| 0.0 | 7.5 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 9.1 | GO:0061061 | muscle structure development(GO:0061061) |
| 0.0 | 2.1 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 16.8 | GO:0006468 | protein phosphorylation(GO:0006468) |
| 0.0 | 7.3 | GO:0030036 | actin cytoskeleton organization(GO:0030036) |
| 0.0 | 0.6 | GO:0045765 | regulation of angiogenesis(GO:0045765) |
| 0.0 | 0.0 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.0 | 2.9 | GO:0045944 | positive regulation of transcription from RNA polymerase II promoter(GO:0045944) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 35.9 | GO:0005869 | dynactin complex(GO:0005869) |
| 2.5 | 15.2 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 1.3 | 24.2 | GO:0031672 | A band(GO:0031672) |
| 1.0 | 5.0 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.8 | 40.8 | GO:0016459 | myosin complex(GO:0016459) |
| 0.5 | 2.1 | GO:0097433 | dense body(GO:0097433) |
| 0.5 | 42.5 | GO:0005884 | actin filament(GO:0005884) |
| 0.5 | 10.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.5 | 6.9 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.5 | 5.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.3 | 2.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.1 | 3.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 1.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 6.6 | GO:0005923 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.1 | 3.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.1 | 13.9 | GO:0015629 | actin cytoskeleton(GO:0015629) |
| 0.0 | 1.0 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 2.0 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 4.6 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 2.1 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 16.1 | GO:0005576 | extracellular region(GO:0005576) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 15.2 | GO:0048763 | ryanodine-sensitive calcium-release channel activity(GO:0005219) calcium-induced calcium release activity(GO:0048763) |
| 1.6 | 17.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 1.1 | 6.7 | GO:0090556 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.7 | 10.3 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.7 | 6.6 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.6 | 2.5 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.5 | 5.2 | GO:0016405 | CoA-ligase activity(GO:0016405) |
| 0.5 | 2.1 | GO:0098973 | structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.5 | 5.0 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.5 | 14.0 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.4 | 9.1 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.4 | 31.2 | GO:0004879 | RNA polymerase II transcription factor activity, ligand-activated sequence-specific DNA binding(GO:0004879) transcription factor activity, direct ligand regulated sequence-specific DNA binding(GO:0098531) |
| 0.4 | 2.5 | GO:0004309 | exopolyphosphatase activity(GO:0004309) |
| 0.4 | 3.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.3 | 40.8 | GO:0003774 | motor activity(GO:0003774) |
| 0.3 | 13.0 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.3 | 14.8 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.3 | 3.8 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.2 | 43.9 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 0.7 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.1 | 6.9 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.1 | 2.0 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.1 | 18.2 | GO:0003779 | actin binding(GO:0003779) |
| 0.1 | 6.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 2.3 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 38.7 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 1.4 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 6.6 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.2 | GO:0051082 | unfolded protein binding(GO:0051082) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 13.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 1.2 | 17.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.5 | 17.8 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.4 | 6.4 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.2 | 3.6 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.1 | 2.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 3.7 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.4 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 1.6 | 13.0 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.8 | 2.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.6 | 14.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.6 | 12.0 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.3 | 19.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 1.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.1 | 7.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.1 | 1.2 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 1.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |