Project
DANIO-CODE
Navigation
Downloads
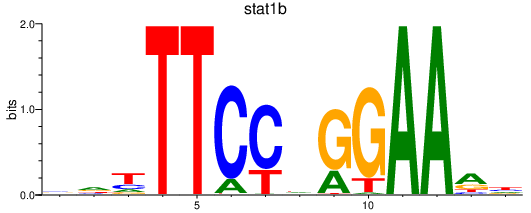
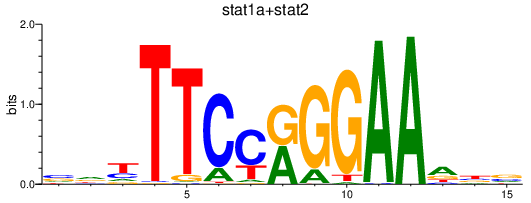
Results for stat1b_stat1a+stat2
Z-value: 0.49
Transcription factors associated with stat1b_stat1a+stat2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
stat1b
|
ENSDARG00000076182 | signal transducer and activator of transcription 1b |
|
stat1a
|
ENSDARG00000006266 | signal transducer and activator of transcription 1a |
|
stat2
|
ENSDARG00000031647 | signal transducer and activator of transcription 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| stat1a | dr10_dc_chr22_+_12773735_12773848 | -0.57 | 2.1e-02 | Click! |
| stat2 | dr10_dc_chr6_-_39162916_39162923 | 0.54 | 3.0e-02 | Click! |
Activity profile of stat1b_stat1a+stat2 motif
Sorted Z-values of stat1b_stat1a+stat2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of stat1b_stat1a+stat2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_15497269 | 0.99 |
ENSDART00000053754
|
has2
|
hyaluronan synthase 2 |
| chr21_+_15774697 | 0.80 |
ENSDART00000122015
|
fam169ab
|
family with sequence similarity 169, member Ab |
| chr21_+_15774822 | 0.73 |
ENSDART00000122015
|
fam169ab
|
family with sequence similarity 169, member Ab |
| chr14_+_6122803 | 0.71 |
ENSDART00000149783
ENSDART00000148461 |
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr11_-_27715592 | 0.70 |
ENSDART00000168338
|
ece1
|
endothelin converting enzyme 1 |
| chr17_+_16038358 | 0.67 |
ENSDART00000155336
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr1_+_41762057 | 0.61 |
ENSDART00000137609
|
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr15_+_24802704 | 0.56 |
ENSDART00000078024
|
crk
|
v-crk avian sarcoma virus CT10 oncogene homolog |
| chr15_+_24802510 | 0.47 |
ENSDART00000078024
|
crk
|
v-crk avian sarcoma virus CT10 oncogene homolog |
| chr17_+_16038103 | 0.46 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr16_-_15496868 | 0.45 |
ENSDART00000053754
|
has2
|
hyaluronan synthase 2 |
| chr8_-_14571519 | 0.45 |
ENSDART00000146175
|
cep350
|
centrosomal protein 350 |
| chr10_-_25448769 | 0.43 |
ENSDART00000140023
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr6_-_54436651 | 0.41 |
ENSDART00000154121
|
sys1
|
Sys1 golgi trafficking protein |
| chr7_+_41033439 | 0.41 |
ENSDART00000016660
|
ENSDARG00000016964
|
ENSDARG00000016964 |
| chr13_-_6875290 | 0.41 |
|
|
|
| chr7_-_28293919 | 0.39 |
ENSDART00000028887
|
tmem9b
|
TMEM9 domain family, member B |
| chr19_+_42936516 | 0.39 |
ENSDART00000165747
|
BX950855.1
|
ENSDARG00000103815 |
| chr6_+_23476669 | 0.39 |
|
|
|
| chr3_-_25144722 | 0.39 |
ENSDART00000055445
|
ddx5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr6_+_41506504 | 0.38 |
ENSDART00000084861
|
cish
|
cytokine inducible SH2-containing protein |
| chr3_+_40147509 | 0.37 |
ENSDART00000017304
|
cpsf4
|
cleavage and polyadenylation specific factor 4 |
| chr5_+_3567992 | 0.36 |
ENSDART00000129329
|
rpain
|
RPA interacting protein |
| chr18_-_26815192 | 0.36 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr6_+_41506681 | 0.36 |
ENSDART00000084861
|
cish
|
cytokine inducible SH2-containing protein |
| chr8_-_7188791 | 0.35 |
ENSDART00000092426
|
grip2a
|
glutamate receptor interacting protein 2a |
| chr23_-_1562603 | 0.34 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr23_-_1562572 | 0.32 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| chr2_+_45119713 | 0.31 |
ENSDART00000075146
|
alg3
|
asparagine-linked glycosylation 3 (alpha-1,3-mannosyltransferase) |
| chr25_-_15176295 | 0.31 |
|
|
|
| chr18_+_6339577 | 0.31 |
ENSDART00000136333
|
wash1
|
WAS protein family homolog 1 |
| chr14_+_6122901 | 0.30 |
ENSDART00000149783
ENSDART00000148461 |
abca1b
|
ATP-binding cassette, sub-family A (ABC1), member 1B |
| chr22_-_3740362 | 0.30 |
|
|
|
| chr3_-_25142506 | 0.30 |
ENSDART00000145315
|
ddx5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr5_+_29193876 | 0.30 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr3_-_25142301 | 0.29 |
ENSDART00000145315
|
ddx5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr19_+_4835526 | 0.29 |
|
|
|
| chr1_+_41762133 | 0.29 |
ENSDART00000137609
|
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr1_+_31609119 | 0.28 |
ENSDART00000135078
|
pudp
|
pseudouridine 5'-phosphatase |
| chr23_-_33631518 | 0.28 |
ENSDART00000018301
|
itga5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr21_+_13291242 | 0.28 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr5_-_18393623 | 0.28 |
ENSDART00000010101
ENSDART00000099434 |
aacs
|
acetoacetyl-CoA synthetase |
| chr13_-_37504883 | 0.27 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr1_+_43352429 | 0.27 |
ENSDART00000124873
|
si:ch73-109d9.4
|
si:ch73-109d9.4 |
| chr19_-_17926712 | 0.26 |
ENSDART00000167592
ENSDART00000162383 |
nkiras1
|
NFKB inhibitor interacting Ras-like 1 |
| chr11_+_7139675 | 0.26 |
ENSDART00000155864
|
CU929070.1
|
ENSDARG00000097452 |
| chr17_+_27045994 | 0.25 |
ENSDART00000178734
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr7_+_22042296 | 0.25 |
ENSDART00000123457
|
TMEM102
|
transmembrane protein 102 |
| chr24_-_19574666 | 0.25 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr15_+_44069851 | 0.25 |
ENSDART00000114352
|
zgc:112998
|
zgc:112998 |
| chr19_+_28363333 | 0.25 |
|
|
|
| chr19_+_32000438 | 0.24 |
ENSDART00000150910
|
gmnn
|
geminin, DNA replication inhibitor |
| chr6_+_12081558 | 0.24 |
ENSDART00000156854
|
si:dkey-276j7.2
|
si:dkey-276j7.2 |
| chr3_-_25142093 | 0.24 |
|
|
|
| chr11_+_19440815 | 0.24 |
ENSDART00000005639
|
thoc7
|
THO complex 7 |
| chr13_-_37482524 | 0.24 |
ENSDART00000140230
|
zgc:123068
|
zgc:123068 |
| chr19_+_4939999 | 0.23 |
ENSDART00000093402
|
cdk12
|
cyclin-dependent kinase 12 |
| chr13_+_22973764 | 0.23 |
ENSDART00000110266
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr8_-_14571436 | 0.23 |
ENSDART00000146175
|
cep350
|
centrosomal protein 350 |
| chr3_-_25142556 | 0.23 |
ENSDART00000145315
|
ddx5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr15_+_20303330 | 0.22 |
ENSDART00000152473
|
spint2
|
serine peptidase inhibitor, Kunitz type, 2 |
| chr18_+_20045544 | 0.22 |
|
|
|
| chr10_+_21477806 | 0.22 |
ENSDART00000142447
|
etf1b
|
eukaryotic translation termination factor 1b |
| chr8_+_47108614 | 0.22 |
ENSDART00000142979
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr20_-_3873641 | 0.22 |
ENSDART00000113132
|
gnpat
|
glyceronephosphate O-acyltransferase |
| chr10_+_21477579 | 0.22 |
ENSDART00000142447
|
etf1b
|
eukaryotic translation termination factor 1b |
| chr20_+_46668743 | 0.22 |
ENSDART00000139051
ENSDART00000161320 |
batf
|
basic leucine zipper transcription factor, ATF-like |
| chr11_-_22143133 | 0.22 |
ENSDART00000159681
|
tfeb
|
transcription factor EB |
| chr20_+_52021000 | 0.21 |
ENSDART00000074330
|
taf1a
|
TATA box binding protein (TBP)-associated factor, RNA polymerase I, A |
| chr21_+_13291305 | 0.21 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr20_-_3220392 | 0.21 |
ENSDART00000123331
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr16_-_47446494 | 0.21 |
ENSDART00000032188
|
si:dkey-256h2.1
|
si:dkey-256h2.1 |
| chr24_-_19574944 | 0.21 |
ENSDART00000135405
|
csrnp1b
|
cysteine-serine-rich nuclear protein 1b |
| chr18_+_20045293 | 0.20 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr2_+_42155813 | 0.20 |
ENSDART00000143562
|
gbp2
|
guanylate binding protein 2 |
| chr3_+_32440178 | 0.20 |
ENSDART00000151025
|
si:ch73-248e21.1
|
si:ch73-248e21.1 |
| chr16_-_2880109 | 0.20 |
ENSDART00000149463
|
hace1
|
HECT domain and ankyrin repeat containing E3 ubiquitin protein ligase 1 |
| chr20_+_14073464 | 0.20 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr10_+_16544000 | 0.19 |
ENSDART00000121864
|
slc27a6
|
solute carrier family 27 (fatty acid transporter), member 6 |
| chr20_-_9135267 | 0.19 |
ENSDART00000125133
|
mysm1
|
Myb-like, SWIRM and MPN domains 1 |
| chr22_+_10683985 | 0.19 |
ENSDART00000063207
ENSDART00000122349 |
hiat1b
|
hippocampus abundant transcript 1b |
| chr24_+_16005004 | 0.19 |
ENSDART00000163086
ENSDART00000152087 |
si:dkey-118j18.2
|
si:dkey-118j18.2 |
| chr14_-_6669062 | 0.19 |
|
|
|
| chr21_+_28710341 | 0.18 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr5_+_36168475 | 0.18 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr8_+_47108500 | 0.18 |
ENSDART00000142979
|
arhgef16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr19_-_25565317 | 0.18 |
ENSDART00000175266
|
ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr15_+_24802607 | 0.18 |
ENSDART00000078024
|
crk
|
v-crk avian sarcoma virus CT10 oncogene homolog |
| chr20_+_27188382 | 0.18 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr24_+_43010 | 0.17 |
ENSDART00000122785
|
zgc:152808
|
zgc:152808 |
| chr5_-_41260390 | 0.17 |
ENSDART00000140154
|
si:dkey-65b12.12
|
si:dkey-65b12.12 |
| chr23_+_44795356 | 0.17 |
ENSDART00000145905
|
eno3
|
enolase 3, (beta, muscle) |
| chr20_-_3220435 | 0.17 |
ENSDART00000123331
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr8_+_19945820 | 0.17 |
ENSDART00000134124
|
znf692
|
zinc finger protein 692 |
| chr21_-_37750338 | 0.17 |
ENSDART00000133585
|
fam114a2
|
family with sequence similarity 114, member A2 |
| chr21_+_13291347 | 0.17 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr4_-_765957 | 0.17 |
ENSDART00000128743
|
tmem214
|
transmembrane protein 214 |
| chr8_-_53165501 | 0.16 |
ENSDART00000135982
|
nr6a1a
|
nuclear receptor subfamily 6, group A, member 1a |
| chr18_-_48329076 | 0.16 |
|
|
|
| chr10_+_17470158 | 0.16 |
|
|
|
| chr18_+_20044862 | 0.16 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr18_+_45669615 | 0.16 |
ENSDART00000150973
|
prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr15_+_29092022 | 0.16 |
ENSDART00000141164
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr5_+_19506512 | 0.16 |
ENSDART00000047841
|
sgsm1a
|
small G protein signaling modulator 1a |
| chr7_-_981576 | 0.16 |
ENSDART00000101799
|
alpk1
|
alpha-kinase 1 |
| chr18_-_26815111 | 0.15 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr16_+_40610589 | 0.15 |
ENSDART00000038294
|
tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr5_+_36054658 | 0.15 |
ENSDART00000131339
|
capns1a
|
calpain, small subunit 1 a |
| chr10_-_25448712 | 0.15 |
ENSDART00000140023
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr11_-_11925832 | 0.15 |
|
|
|
| chr20_-_3220475 | 0.15 |
ENSDART00000123331
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr24_-_5904097 | 0.15 |
ENSDART00000007373
ENSDART00000135124 |
acbd5a
|
acyl-CoA binding domain containing 5a |
| chr19_+_30212276 | 0.15 |
|
|
|
| chr9_-_12472519 | 0.14 |
ENSDART00000102434
|
ehhadh
|
enoyl-CoA, hydratase/3-hydroxyacyl CoA dehydrogenase |
| chr25_+_32770866 | 0.14 |
ENSDART00000059800
|
lactb
|
lactamase, beta |
| chr16_-_55211053 | 0.14 |
ENSDART00000156533
|
kdf1a
|
keratinocyte differentiation factor 1a |
| chr23_-_25853364 | 0.14 |
ENSDART00000110670
|
ENSDARG00000078574
|
ENSDARG00000078574 |
| chr8_-_16577737 | 0.14 |
ENSDART00000139038
|
CU467861.1
|
ENSDARG00000092337 |
| chr19_-_11001651 | 0.14 |
ENSDART00000165630
|
ecm1a
|
extracellular matrix protein 1a |
| chr11_+_12662507 | 0.14 |
ENSDART00000135761
|
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr25_-_35094522 | 0.14 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr16_-_14973966 | 0.14 |
|
|
|
| chr10_+_17470092 | 0.14 |
|
|
|
| chr15_+_29091983 | 0.13 |
ENSDART00000141164
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr2_+_4366348 | 0.13 |
ENSDART00000166476
|
gata6
|
GATA binding protein 6 |
| chr7_+_41033650 | 0.13 |
ENSDART00000016660
|
ENSDARG00000016964
|
ENSDARG00000016964 |
| chr23_-_25853220 | 0.13 |
ENSDART00000110670
|
ENSDARG00000078574
|
ENSDARG00000078574 |
| chr6_-_39313358 | 0.13 |
|
|
|
| chr3_+_36504142 | 0.13 |
ENSDART00000170013
|
gspt1l
|
G1 to S phase transition 1, like |
| chr22_-_16264430 | 0.13 |
ENSDART00000021666
|
rtca
|
RNA 3'-terminal phosphate cyclase |
| chr2_+_26632673 | 0.13 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr15_-_5811394 | 0.13 |
ENSDART00000158522
|
hnrnpl2
|
heterogeneous nuclear ribonucleoprotein L2 |
| chr3_+_26092485 | 0.12 |
ENSDART00000128284
|
nudt9
|
nudix (nucleoside diphosphate linked moiety X)-type motif 9 |
| chr20_-_29780948 | 0.12 |
ENSDART00000131219
ENSDART00000015928 |
si:ch211-195d17.2
|
si:ch211-195d17.2 |
| chr23_+_39569262 | 0.12 |
ENSDART00000031616
|
mical1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr7_-_981435 | 0.12 |
ENSDART00000101799
|
alpk1
|
alpha-kinase 1 |
| chr8_+_23131037 | 0.12 |
ENSDART00000036801
|
slc17a9a
|
solute carrier family 17 (vesicular nucleotide transporter), member 9a |
| chr14_+_38506051 | 0.12 |
ENSDART00000144599
|
hnrnpa0b
|
heterogeneous nuclear ribonucleoprotein A0b |
| chr7_+_26274252 | 0.12 |
ENSDART00000164824
|
tnk1
|
tyrosine kinase, non-receptor, 1 |
| chr15_+_24802238 | 0.12 |
ENSDART00000078024
|
crk
|
v-crk avian sarcoma virus CT10 oncogene homolog |
| chr16_-_42202425 | 0.12 |
ENSDART00000038748
|
dyrk1b
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr20_-_3220106 | 0.12 |
ENSDART00000123331
|
spint1b
|
serine peptidase inhibitor, Kunitz type 1 b |
| chr17_+_27046020 | 0.11 |
ENSDART00000178734
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr1_-_9789386 | 0.11 |
ENSDART00000040116
|
tnrc5
|
trinucleotide repeat containing 5 |
| chr2_-_26934833 | 0.11 |
ENSDART00000025120
|
ndc1
|
NDC1 transmembrane nucleoporin |
| chr2_+_35750334 | 0.11 |
ENSDART00000052666
|
rasal2
|
RAS protein activator like 2 |
| chr15_-_23786063 | 0.11 |
ENSDART00000109318
|
zc3h4
|
zinc finger CCCH-type containing 4 |
| chr5_-_18393705 | 0.11 |
ENSDART00000010101
ENSDART00000099434 |
aacs
|
acetoacetyl-CoA synthetase |
| chr18_-_16964811 | 0.11 |
ENSDART00000100126
|
akip1
|
A kinase (PRKA) interacting protein 1 |
| chr12_-_46943717 | 0.11 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr21_+_10689336 | 0.11 |
ENSDART00000146248
|
znf532
|
zinc finger protein 532 |
| chr21_+_28710269 | 0.11 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr22_-_26112676 | 0.11 |
|
|
|
| chr18_+_36085169 | 0.11 |
ENSDART00000059347
|
bckdha
|
branched chain keto acid dehydrogenase E1, alpha polypeptide |
| chr6_-_39313081 | 0.11 |
|
|
|
| chr11_-_18637950 | 0.11 |
ENSDART00000156276
|
pfkfb2b
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2b |
| chr21_-_40759739 | 0.10 |
ENSDART00000075808
|
apbb3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr19_+_7839888 | 0.10 |
|
|
|
| chr25_-_36691657 | 0.10 |
ENSDART00000153789
|
rfwd3
|
ring finger and WD repeat domain 3 |
| chr5_+_67157818 | 0.10 |
ENSDART00000162301
ENSDART00000159386 |
si:ch211-271b14.1
|
si:ch211-271b14.1 |
| chr13_-_49920975 | 0.10 |
ENSDART00000165066
|
eif2ak2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
| chr5_+_56988663 | 0.10 |
ENSDART00000074268
|
zgc:153929
|
zgc:153929 |
| chr8_+_7625846 | 0.10 |
ENSDART00000175045
|
CABZ01078218.1
|
ENSDARG00000107577 |
| chr22_-_10744909 | 0.10 |
ENSDART00000081156
|
timm13
|
translocase of inner mitochondrial membrane 13 homolog (yeast) |
| chr15_-_23550450 | 0.10 |
ENSDART00000152706
|
nlrx1
|
NLR family member X1 |
| chr3_-_40791555 | 0.10 |
ENSDART00000055201
|
foxk1
|
forkhead box K1 |
| chr14_-_32404076 | 0.10 |
ENSDART00000075617
|
sox3
|
SRY (sex determining region Y)-box 3 |
| chr10_+_24530670 | 0.10 |
|
|
|
| chr7_-_38363533 | 0.10 |
ENSDART00000173861
|
c1qtnf4
|
C1q and TNF related 4 |
| chr10_-_17211813 | 0.10 |
ENSDART00000020122
|
ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
| chr2_-_7772932 | 0.09 |
ENSDART00000160053
|
ripk2
|
receptor-interacting serine-threonine kinase 2 |
| chr5_-_56953587 | 0.09 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr5_-_51568854 | 0.09 |
ENSDART00000163973
|
lnpep
|
leucyl/cystinyl aminopeptidase |
| chr22_+_4035577 | 0.09 |
ENSDART00000170620
|
ctxn1
|
cortexin 1 |
| chr3_+_24381364 | 0.09 |
ENSDART00000133898
|
dnal4a
|
dynein, axonemal, light chain 4a |
| chr9_+_18565169 | 0.09 |
|
|
|
| chr25_+_32770756 | 0.09 |
ENSDART00000059800
|
lactb
|
lactamase, beta |
| chr20_-_37910887 | 0.09 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr12_+_47828020 | 0.09 |
ENSDART00000112010
|
lzts2b
|
leucine zipper, putative tumor suppressor 2b |
| chr3_-_13449591 | 0.09 |
ENSDART00000166639
|
tufm
|
Tu translation elongation factor, mitochondrial |
| chr11_-_11925801 | 0.09 |
|
|
|
| chr20_+_14073428 | 0.09 |
ENSDART00000049864
|
rd3
|
retinal degeneration 3 |
| chr15_+_12504537 | 0.08 |
ENSDART00000168011
|
tmprss4a
|
transmembrane protease, serine 4a |
| chr4_+_3442322 | 0.08 |
ENSDART00000058277
|
znf800b
|
zinc finger protein 800b |
| chr13_-_37482466 | 0.08 |
ENSDART00000147934
|
zgc:123068
|
zgc:123068 |
| chr1_-_22679567 | 0.08 |
ENSDART00000126785
|
pds5a
|
PDS5 cohesin associated factor A |
| chr11_+_12662882 | 0.08 |
ENSDART00000122812
|
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr24_+_42964 | 0.08 |
ENSDART00000122785
|
zgc:152808
|
zgc:152808 |
| chr18_-_26815261 | 0.08 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr6_-_43297126 | 0.08 |
ENSDART00000154170
|
frmd4ba
|
FERM domain containing 4Ba |
| chr7_+_17701669 | 0.08 |
ENSDART00000113120
|
taf6l
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chr25_+_32770967 | 0.08 |
ENSDART00000059800
|
lactb
|
lactamase, beta |
| chr7_-_45747162 | 0.08 |
ENSDART00000172591
|
zgc:162297
|
zgc:162297 |
| chr3_+_703484 | 0.08 |
ENSDART00000134971
|
zgc:194659
|
zgc:194659 |
| chr15_+_40228295 | 0.08 |
ENSDART00000111018
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr5_-_13844232 | 0.08 |
|
|
|
| chr18_+_20045221 | 0.08 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr7_-_67741663 | 0.08 |
|
|
|
| chr21_+_28710424 | 0.08 |
ENSDART00000014058
|
zgc:100829
|
zgc:100829 |
| chr6_-_39313219 | 0.07 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0046379 | renal water homeostasis(GO:0003091) renal water transport(GO:0003097) extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.2 | 0.9 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 0.4 | GO:0002793 | positive regulation of peptide secretion(GO:0002793) positive regulation of insulin secretion(GO:0032024) positive regulation of hormone secretion(GO:0046887) positive regulation of peptide hormone secretion(GO:0090277) |
| 0.1 | 0.6 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.1 | 0.6 | GO:0033700 | phospholipid efflux(GO:0033700) |
| 0.1 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.2 | GO:0043368 | T-helper cell lineage commitment(GO:0002295) T cell lineage commitment(GO:0002360) alpha-beta T cell lineage commitment(GO:0002363) positive T cell selection(GO:0043368) CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) T cell selection(GO:0045058) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 | 0.2 | GO:2000378 | negative regulation of reactive oxygen species metabolic process(GO:2000378) |
| 0.1 | 0.7 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.1 | 0.3 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 0.4 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.6 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.2 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.0 | 1.3 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.7 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.4 | GO:0098787 | mRNA cleavage involved in mRNA processing(GO:0098787) pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.1 | GO:0010481 | keratinocyte development(GO:0003334) epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.6 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.1 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.0 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.2 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.3 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.0 | 0.1 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.2 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.2 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.6 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.2 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.4 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.2 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.3 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) nuclear cyclin-dependent protein kinase holoenzyme complex(GO:0019908) |
| 0.0 | 0.1 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 0.5 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.1 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.0 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.2 | 0.5 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.1 | 0.6 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.3 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.2 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 0.6 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.2 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.4 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.2 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0004044 | amidophosphoribosyltransferase activity(GO:0004044) |
| 0.0 | 0.2 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.3 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.2 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.0 | 0.2 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.0 | 1.2 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.0 | 0.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.4 | GO:0016878 | acid-thiol ligase activity(GO:0016878) |
| 0.0 | 0.1 | GO:0019705 | protein-cysteine S-myristoyltransferase activity(GO:0019705) |
| 0.0 | 0.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.2 | GO:0031267 | small GTPase binding(GO:0031267) |
| 0.0 | 0.0 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.2 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.1 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.0 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.7 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.4 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 0.6 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.4 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.5 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |