Project
DANIO-CODE
Navigation
Downloads
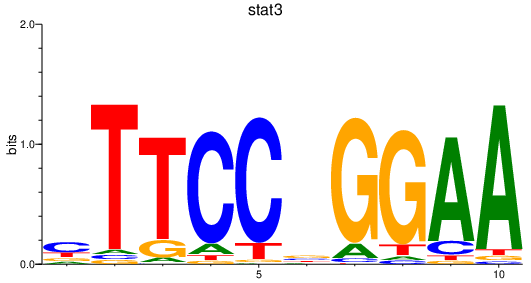
Results for stat3
Z-value: 0.67
Transcription factors associated with stat3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
stat3
|
ENSDARG00000022712 | signal transducer and activator of transcription 3 (acute-phase response factor) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| stat3 | dr10_dc_chr3_+_16880842_16880919 | -0.49 | 5.5e-02 | Click! |
Activity profile of stat3 motif
Sorted Z-values of stat3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of stat3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_41506220 | 2.00 |
ENSDART00000136538
|
cish
|
cytokine inducible SH2-containing protein |
| chr6_+_41506350 | 1.75 |
ENSDART00000136538
|
cish
|
cytokine inducible SH2-containing protein |
| chr6_+_41506446 | 1.15 |
ENSDART00000140108
|
cish
|
cytokine inducible SH2-containing protein |
| chr20_+_27188382 | 0.77 |
ENSDART00000062094
|
tmem251
|
transmembrane protein 251 |
| chr6_+_41506504 | 0.68 |
ENSDART00000084861
|
cish
|
cytokine inducible SH2-containing protein |
| chr10_+_40362746 | 0.64 |
ENSDART00000062795
ENSDART00000113582 |
git2b
|
G protein-coupled receptor kinase interacting ArfGAP 2b |
| KN149738v1_-_5273 | 0.63 |
|
|
|
| chr13_+_22589282 | 0.61 |
ENSDART00000057672
|
nfkb2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr20_+_19094480 | 0.58 |
ENSDART00000152380
|
tdh
|
L-threonine dehydrogenase |
| chr3_+_26682186 | 0.57 |
ENSDART00000055537
|
socs1a
|
suppressor of cytokine signaling 1a |
| chr19_+_48144910 | 0.55 |
ENSDART00000149705
|
cdca8
|
cell division cycle associated 8 |
| chr21_+_25740782 | 0.55 |
ENSDART00000021620
|
cldnd
|
claudin d |
| KN149738v1_-_5192 | 0.54 |
|
|
|
| chr24_+_28571936 | 0.52 |
ENSDART00000153933
|
CR933730.1
|
ENSDARG00000097841 |
| chr9_-_43842377 | 0.52 |
ENSDART00000023684
|
cwc22
|
CWC22 spliceosome-associated protein homolog (S. cerevisiae) |
| chr6_+_12999546 | 0.50 |
ENSDART00000089725
|
ino80db
|
INO80 complex subunit Db |
| chr5_+_36168475 | 0.49 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr6_+_12999630 | 0.45 |
ENSDART00000089725
|
ino80db
|
INO80 complex subunit Db |
| chr20_+_46668743 | 0.44 |
ENSDART00000139051
ENSDART00000161320 |
batf
|
basic leucine zipper transcription factor, ATF-like |
| chr20_-_26161022 | 0.44 |
ENSDART00000151950
|
serac1
|
serine active site containing 1 |
| chr10_-_7813660 | 0.43 |
ENSDART00000158559
|
si:ch73-340n8.4
|
si:ch73-340n8.4 |
| chr4_-_1968907 | 0.42 |
ENSDART00000067433
|
ube2nb
|
ubiquitin-conjugating enzyme E2Nb |
| chr12_-_35467285 | 0.41 |
ENSDART00000160336
|
sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr13_-_37504883 | 0.41 |
ENSDART00000108855
|
si:dkey-188i13.7
|
si:dkey-188i13.7 |
| chr21_+_39649559 | 0.41 |
ENSDART00000100240
|
rnmtl1b
|
RNA methyltransferase like 1b |
| chr22_+_2014357 | 0.40 |
ENSDART00000165168
|
znf1157
|
zinc finger protein 1157 |
| chr8_+_27788830 | 0.40 |
ENSDART00000078509
|
capza1b
|
capping protein (actin filament) muscle Z-line, alpha 1b |
| chr8_-_19187191 | 0.40 |
|
|
|
| chr19_-_1376498 | 0.40 |
ENSDART00000158429
|
tmem42b
|
transmembrane protein 42b |
| chr12_-_4597113 | 0.38 |
ENSDART00000110514
|
prr12a
|
proline rich 12a |
| chr6_-_3837294 | 0.38 |
ENSDART00000059212
|
unc50
|
unc-50 homolog (C. elegans) |
| chr9_-_12687806 | 0.38 |
ENSDART00000058565
|
pttg1ipb
|
PTTG1 interacting protein b |
| chr1_+_40758474 | 0.37 |
ENSDART00000139521
|
si:ch211-89o9.4
|
si:ch211-89o9.4 |
| chr3_+_35412519 | 0.36 |
ENSDART00000084549
|
rpusd1
|
RNA pseudouridylate synthase domain containing 1 |
| chr23_-_1562572 | 0.36 |
ENSDART00000013635
|
fbxo30b
|
F-box protein 30b |
| KN150339v1_-_47655 | 0.36 |
|
|
|
| chr3_+_1450673 | 0.36 |
ENSDART00000161945
|
rrp7a
|
ribosomal RNA processing 7 homolog A |
| chr23_+_20592121 | 0.36 |
ENSDART00000114246
|
adnpb
|
activity-dependent neuroprotector homeobox b |
| chr8_+_27788761 | 0.35 |
ENSDART00000170037
|
capza1b
|
capping protein (actin filament) muscle Z-line, alpha 1b |
| chr5_-_41260390 | 0.35 |
ENSDART00000140154
|
si:dkey-65b12.12
|
si:dkey-65b12.12 |
| chr12_-_4584327 | 0.35 |
|
|
|
| chr15_-_1519615 | 0.35 |
ENSDART00000129356
|
si:dkeyp-97b10.3
|
si:dkeyp-97b10.3 |
| chr6_+_40469996 | 0.34 |
ENSDART00000103868
|
zgc:152986
|
zgc:152986 |
| chr3_-_52406525 | 0.34 |
ENSDART00000138018
|
zgc:113210
|
zgc:113210 |
| chr19_-_48144752 | 0.34 |
ENSDART00000158979
|
c19h1orf109
|
c19h1orf109 homolog (H. sapiens) |
| chr5_+_3567992 | 0.34 |
ENSDART00000129329
|
rpain
|
RPA interacting protein |
| chr24_+_14792755 | 0.33 |
ENSDART00000091735
|
dok6
|
docking protein 6 |
| chr3_-_40791035 | 0.33 |
ENSDART00000055201
|
foxk1
|
forkhead box K1 |
| chr8_+_52528853 | 0.33 |
ENSDART00000163668
|
si:ch1073-392o20.2
|
si:ch1073-392o20.2 |
| chr3_-_35411760 | 0.33 |
ENSDART00000022147
|
ndufab1b
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1b |
| chr19_-_33356861 | 0.31 |
ENSDART00000137611
|
azin1b
|
antizyme inhibitor 1b |
| chr21_-_43332911 | 0.31 |
ENSDART00000114955
|
sowahaa
|
sosondowah ankyrin repeat domain family member Aa |
| chr24_-_5903682 | 0.31 |
ENSDART00000007373
ENSDART00000135124 |
acbd5a
|
acyl-CoA binding domain containing 5a |
| chr13_+_9510368 | 0.30 |
ENSDART00000140504
|
prdx3
|
peroxiredoxin 3 |
| chr19_+_14132374 | 0.30 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| chr17_+_27046020 | 0.29 |
ENSDART00000178734
|
rps6ka1
|
ribosomal protein S6 kinase a, polypeptide 1 |
| chr24_+_5176362 | 0.29 |
ENSDART00000155926
|
si:ch73-206p6.1
|
si:ch73-206p6.1 |
| chr18_+_20236794 | 0.29 |
ENSDART00000045679
|
tle3a
|
transducin-like enhancer of split 3a |
| chr3_-_35411825 | 0.29 |
ENSDART00000022147
|
ndufab1b
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1b |
| chr17_-_9794176 | 0.29 |
|
|
|
| chr19_-_48144715 | 0.29 |
ENSDART00000158979
|
c19h1orf109
|
c19h1orf109 homolog (H. sapiens) |
| chr7_+_28630711 | 0.28 |
ENSDART00000113998
|
dok4
|
docking protein 4 |
| chr3_-_40790889 | 0.28 |
|
|
|
| chr25_+_33118422 | 0.28 |
|
|
|
| chr2_-_7772932 | 0.27 |
ENSDART00000160053
|
ripk2
|
receptor-interacting serine-threonine kinase 2 |
| chr18_+_18992998 | 0.27 |
ENSDART00000006300
|
hacd3
|
3-hydroxyacyl-CoA dehydratase 3 |
| chr7_+_22042296 | 0.27 |
ENSDART00000123457
|
TMEM102
|
transmembrane protein 102 |
| chr20_-_37910887 | 0.27 |
ENSDART00000147529
|
batf3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr7_+_28630661 | 0.27 |
ENSDART00000113998
|
dok4
|
docking protein 4 |
| chr3_+_1451017 | 0.27 |
ENSDART00000161945
|
rrp7a
|
ribosomal RNA processing 7 homolog A |
| chr13_-_50053533 | 0.26 |
ENSDART00000098209
|
sirt1
|
sirtuin 1 |
| chr19_+_5501487 | 0.26 |
ENSDART00000132874
|
eif1b
|
eukaryotic translation initiation factor 1B |
| chr6_-_3837266 | 0.25 |
ENSDART00000059212
|
unc50
|
unc-50 homolog (C. elegans) |
| chr9_+_44503204 | 0.25 |
ENSDART00000147990
|
ssfa2
|
sperm specific antigen 2 |
| chr6_+_12999420 | 0.25 |
ENSDART00000089725
|
ino80db
|
INO80 complex subunit Db |
| chr10_+_24530670 | 0.25 |
|
|
|
| chr18_+_14715573 | 0.24 |
ENSDART00000108469
|
spata2l
|
spermatogenesis associated 2-like |
| chr7_-_18916067 | 0.24 |
ENSDART00000112447
|
il13ra1
|
interleukin 13 receptor, alpha 1 |
| chr5_+_26888744 | 0.24 |
ENSDART00000123635
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr10_-_7813594 | 0.24 |
ENSDART00000158559
|
si:ch73-340n8.4
|
si:ch73-340n8.4 |
| chr11_-_27491564 | 0.24 |
ENSDART00000021949
|
fam120a
|
family with sequence similarity 120A |
| chr14_+_30390987 | 0.23 |
|
|
|
| chr20_-_25652858 | 0.23 |
ENSDART00000003666
ENSDART00000122351 ENSDART00000063081 ENSDART00000153282 |
cyp2ad3
|
cytochrome P450, family 2, subfamily AD, polypeptide 3 |
| chr9_-_21427035 | 0.23 |
ENSDART00000146764
|
cox17
|
COX17 cytochrome c oxidase copper chaperone |
| chr8_+_19516856 | 0.22 |
ENSDART00000123104
|
notch2
|
notch 2 |
| chr25_-_7539433 | 0.22 |
ENSDART00000131415
|
arpp19b
|
cAMP-regulated phosphoprotein 19b |
| chr3_+_17901295 | 0.22 |
ENSDART00000035531
|
mettl26
|
methyltransferase like 26 |
| chr2_+_37855019 | 0.21 |
ENSDART00000113337
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr24_-_36307897 | 0.21 |
ENSDART00000154395
|
esco1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr13_+_22973830 | 0.21 |
ENSDART00000110266
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr19_-_48144624 | 0.21 |
ENSDART00000158979
|
c19h1orf109
|
c19h1orf109 homolog (H. sapiens) |
| KN150170v1_+_28652 | 0.21 |
|
|
|
| chr13_+_22973764 | 0.21 |
ENSDART00000110266
|
pik3ap1
|
phosphoinositide-3-kinase adaptor protein 1 |
| chr5_+_36295353 | 0.20 |
ENSDART00000135776
|
srsf7a
|
serine/arginine-rich splicing factor 7a |
| chr12_+_18914438 | 0.20 |
ENSDART00000153086
|
kctd17
|
potassium channel tetramerization domain containing 17 |
| chr21_-_43462380 | 0.20 |
ENSDART00000085039
|
stk26
|
serine/threonine protein kinase 26 |
| chr4_+_74816506 | 0.20 |
|
|
|
| chr1_-_15814788 | 0.20 |
ENSDART00000169081
|
mtmr7b
|
myotubularin related protein 7b |
| chr8_-_20882626 | 0.20 |
ENSDART00000025356
|
haus5
|
HAUS augmin-like complex, subunit 5 |
| chr12_-_4597080 | 0.20 |
ENSDART00000110514
|
prr12a
|
proline rich 12a |
| chr10_-_16910230 | 0.20 |
ENSDART00000171755
ENSDART00000170953 |
stoml2
|
stomatin (EPB72)-like 2 |
| chr8_-_41511380 | 0.19 |
ENSDART00000019858
|
golga1
|
golgin A1 |
| chr5_-_56254422 | 0.19 |
ENSDART00000014028
|
ppm1db
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Db |
| chr5_-_12086676 | 0.19 |
ENSDART00000081494
ENSDART00000162780 |
vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr23_-_33728344 | 0.19 |
ENSDART00000146180
|
csrnp2
|
cysteine-serine-rich nuclear protein 2 |
| chr22_+_2014232 | 0.19 |
ENSDART00000165168
|
znf1157
|
zinc finger protein 1157 |
| chr3_+_4591775 | 0.19 |
ENSDART00000088610
|
rangap1a
|
RAN GTPase activating protein 1a |
| chr3_-_40791490 | 0.19 |
ENSDART00000055201
|
foxk1
|
forkhead box K1 |
| chr7_+_41033728 | 0.19 |
ENSDART00000016660
|
ENSDARG00000016964
|
ENSDARG00000016964 |
| chr21_+_43333462 | 0.19 |
ENSDART00000109620
|
sept8a
|
septin 8a |
| chr1_+_40057806 | 0.19 |
ENSDART00000175582
|
gpr78a
|
G protein-coupled receptor 78a |
| chr6_+_40575726 | 0.18 |
ENSDART00000004075
|
uqcc2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| chr21_+_13291305 | 0.18 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr23_-_35997065 | 0.18 |
ENSDART00000153910
|
BX005254.4
|
ENSDARG00000097150 |
| chr3_-_30927275 | 0.18 |
ENSDART00000112266
|
armc5
|
armadillo repeat containing 5 |
| chr1_-_51100156 | 0.18 |
ENSDART00000109640
|
junba
|
jun B proto-oncogene a |
| chr5_+_67157818 | 0.18 |
ENSDART00000162301
ENSDART00000159386 |
si:ch211-271b14.1
|
si:ch211-271b14.1 |
| chr22_-_817086 | 0.18 |
ENSDART00000105873
|
cry4
|
cryptochrome circadian clock 4 |
| chr23_-_20592068 | 0.18 |
ENSDART00000153767
|
CR352228.1
|
ENSDARG00000097895 |
| chr10_-_16910257 | 0.17 |
ENSDART00000171755
ENSDART00000170953 |
stoml2
|
stomatin (EPB72)-like 2 |
| chr22_-_26112676 | 0.17 |
|
|
|
| chr3_+_43361636 | 0.17 |
ENSDART00000161277
|
nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr2_+_37854726 | 0.17 |
ENSDART00000113337
|
parp2
|
poly (ADP-ribose) polymerase 2 |
| chr17_+_16038358 | 0.17 |
ENSDART00000155336
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr13_-_25418278 | 0.17 |
ENSDART00000077655
ENSDART00000168099 ENSDART00000135788 |
mcmbp
|
minichromosome maintenance complex binding protein |
| chr6_+_20008821 | 0.17 |
ENSDART00000151205
|
aldh1l1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr24_+_39139519 | 0.17 |
ENSDART00000085565
|
capn15
|
calpain 15 |
| chr10_-_25448712 | 0.17 |
ENSDART00000140023
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr12_+_3632548 | 0.16 |
|
|
|
| chr8_-_31691921 | 0.16 |
ENSDART00000141799
|
fbxo4
|
F-box protein 4 |
| chr19_+_14132473 | 0.16 |
ENSDART00000164696
|
tmem222b
|
transmembrane protein 222b |
| KN149738v1_-_5245 | 0.16 |
|
|
|
| chr23_-_19789409 | 0.16 |
ENSDART00000131860
|
rpl10
|
ribosomal protein L10 |
| chr9_+_12473243 | 0.16 |
ENSDART00000132709
|
tmem41aa
|
transmembrane protein 41aa |
| chr10_+_17634410 | 0.16 |
ENSDART00000168635
|
KCNN2
|
potassium calcium-activated channel subfamily N member 2 |
| chr6_+_40575775 | 0.16 |
ENSDART00000004075
|
uqcc2
|
ubiquinol-cytochrome c reductase complex assembly factor 2 |
| KN149738v1_-_5321 | 0.16 |
|
|
|
| chr18_+_20045506 | 0.16 |
|
|
|
| chr21_+_13291347 | 0.16 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr1_-_54331157 | 0.16 |
ENSDART00000083572
|
zgc:136864
|
zgc:136864 |
| chr3_-_40791555 | 0.16 |
ENSDART00000055201
|
foxk1
|
forkhead box K1 |
| chr20_+_19093849 | 0.16 |
ENSDART00000025509
|
tdh
|
L-threonine dehydrogenase |
| KN150339v1_-_34277 | 0.16 |
|
|
|
| chr3_+_17901095 | 0.15 |
ENSDART00000035531
|
mettl26
|
methyltransferase like 26 |
| chr5_-_56254353 | 0.15 |
ENSDART00000014028
|
ppm1db
|
protein phosphatase, Mg2+/Mn2+ dependent, 1Db |
| chr21_-_3462128 | 0.15 |
ENSDART00000139194
|
dym
|
dymeclin |
| chr18_+_22228682 | 0.15 |
ENSDART00000165464
|
fam65a
|
family with sequence similarity 65, member A |
| chr6_+_36843778 | 0.15 |
ENSDART00000104157
|
tmem45a
|
transmembrane protein 45a |
| chr10_-_25448769 | 0.14 |
ENSDART00000140023
|
grik1a
|
glutamate receptor, ionotropic, kainate 1a |
| chr24_-_5904097 | 0.14 |
ENSDART00000007373
ENSDART00000135124 |
acbd5a
|
acyl-CoA binding domain containing 5a |
| chr5_+_26888419 | 0.14 |
ENSDART00000123635
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr21_-_2191980 | 0.14 |
ENSDART00000159315
|
si:dkey-50i6.5
|
si:dkey-50i6.5 |
| chr5_+_19944746 | 0.14 |
ENSDART00000124545
|
sart3
|
squamous cell carcinoma antigen recognized by T cells 3 |
| chr3_+_16772351 | 0.14 |
ENSDART00000164895
|
atp6v0a1a
|
ATPase, H+ transporting, lysosomal V0 subunit a1a |
| chr16_+_31966419 | 0.14 |
ENSDART00000131661
|
rps9
|
ribosomal protein S9 |
| chr3_+_43361682 | 0.14 |
ENSDART00000168784
|
nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr6_-_34237257 | 0.13 |
ENSDART00000102391
|
dmrt2b
|
doublesex and mab-3 related transcription factor 2b |
| chr22_+_2014149 | 0.13 |
ENSDART00000165168
|
znf1157
|
zinc finger protein 1157 |
| chr21_+_3017338 | 0.13 |
|
|
|
| chr14_-_1186603 | 0.13 |
|
|
|
| chr11_+_15905877 | 0.13 |
ENSDART00000111284
|
agtrap
|
angiotensin II receptor-associated protein |
| chr19_-_17926712 | 0.13 |
ENSDART00000167592
ENSDART00000162383 |
nkiras1
|
NFKB inhibitor interacting Ras-like 1 |
| chr15_+_40228295 | 0.13 |
ENSDART00000111018
|
ngef
|
neuronal guanine nucleotide exchange factor |
| chr8_+_39964882 | 0.13 |
ENSDART00000134452
|
ggt5a
|
gamma-glutamyltransferase 5a |
| chr24_-_26339981 | 0.13 |
ENSDART00000135496
|
eif5a
|
eukaryotic translation initiation factor 5A |
| chr11_+_12662882 | 0.12 |
ENSDART00000122812
|
ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr21_+_13291242 | 0.12 |
ENSDART00000134347
|
zgc:113162
|
zgc:113162 |
| chr8_-_25547089 | 0.12 |
ENSDART00000078022
|
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr14_+_11151485 | 0.12 |
ENSDART00000169202
|
si:ch211-153b23.5
|
si:ch211-153b23.5 |
| chr3_-_13311188 | 0.12 |
ENSDART00000080807
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr14_+_7626683 | 0.12 |
ENSDART00000171657
|
cxxc5b
|
CXXC finger protein 5b |
| chr15_+_3136812 | 0.11 |
ENSDART00000130968
|
rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr18_+_20045544 | 0.11 |
|
|
|
| chr17_+_16038103 | 0.11 |
ENSDART00000155005
|
si:ch73-204p21.2
|
si:ch73-204p21.2 |
| chr20_+_25652930 | 0.11 |
ENSDART00000162080
|
cyp2v1
|
cytochrome P450, family 2, subfamily V, polypeptide 1 |
| chr10_-_17211813 | 0.11 |
ENSDART00000020122
|
ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
| chr6_-_40924038 | 0.11 |
ENSDART00000076097
|
sfi1
|
SFI1 centrin binding protein |
| chr24_-_18302095 | 0.11 |
ENSDART00000144203
ENSDART00000146024 |
sec61g
|
Sec61 translocon gamma subunit |
| chr17_-_11175808 | 0.10 |
ENSDART00000015418
|
irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr18_-_26815192 | 0.10 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr15_-_5811394 | 0.10 |
ENSDART00000158522
|
hnrnpl2
|
heterogeneous nuclear ribonucleoprotein L2 |
| chr21_+_8249235 | 0.10 |
ENSDART00000129749
|
psmb7
|
proteasome subunit beta 7 |
| chr1_+_43352429 | 0.10 |
ENSDART00000124873
|
si:ch73-109d9.4
|
si:ch73-109d9.4 |
| chr18_+_20045293 | 0.10 |
ENSDART00000139441
|
morf4l1
|
mortality factor 4 like 1 |
| chr18_-_26815261 | 0.09 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr3_-_44466637 | 0.09 |
ENSDART00000169923
|
ndel1b
|
nudE neurodevelopment protein 1-like 1b |
| chr9_-_21042485 | 0.09 |
ENSDART00000028247
|
gdap2
|
ganglioside induced differentiation associated protein 2 |
| chr6_-_40575618 | 0.09 |
ENSDART00000155291
|
tomm6
|
translocase of outer mitochondrial membrane 6 homolog (yeast) |
| chr11_+_4006862 | 0.09 |
ENSDART00000041417
|
camk1b
|
calcium/calmodulin-dependent protein kinase Ib |
| chr18_-_26815111 | 0.08 |
ENSDART00000008013
|
sec11a
|
SEC11 homolog A, signal peptidase complex subunit |
| chr7_+_52984549 | 0.08 |
ENSDART00000169830
|
trip4
|
thyroid hormone receptor interactor 4 |
| chr12_-_1435770 | 0.08 |
|
|
|
| chr3_+_17900981 | 0.08 |
ENSDART00000035531
|
mettl26
|
methyltransferase like 26 |
| chr3_+_32700750 | 0.08 |
ENSDART00000139410
|
cd2bp2
|
CD2 (cytoplasmic tail) binding protein 2 |
| chr6_-_40924459 | 0.08 |
ENSDART00000076097
|
sfi1
|
SFI1 centrin binding protein |
| chr23_-_5849341 | 0.07 |
ENSDART00000019455
|
csrp1a
|
cysteine and glycine-rich protein 1a |
| chr21_+_3017623 | 0.07 |
|
|
|
| chr16_+_51597409 | 0.07 |
|
|
|
| chr2_-_37854616 | 0.07 |
ENSDART00000171409
|
mettl17
|
methyltransferase like 17 |
| chr24_-_39970897 | 0.07 |
ENSDART00000134251
|
slc12a7b
|
solute carrier family 12 (potassium/chloride transporter), member 7b |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.6 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.2 | 0.6 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.1 | 0.7 | GO:0006567 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.1 | 0.4 | GO:0042092 | T-helper cell lineage commitment(GO:0002295) T cell lineage commitment(GO:0002360) alpha-beta T cell lineage commitment(GO:0002363) type 2 immune response(GO:0042092) positive T cell selection(GO:0043368) CD4-positive or CD8-positive, alpha-beta T cell lineage commitment(GO:0043369) CD4-positive, alpha-beta T cell lineage commitment(GO:0043373) T cell selection(GO:0045058) T-helper 2 cell differentiation(GO:0045064) T-helper 17 type immune response(GO:0072538) T-helper 17 cell differentiation(GO:0072539) T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 | 0.3 | GO:1902047 | polyamine transport(GO:0015846) polyamine transmembrane transport(GO:1902047) regulation of polyamine transmembrane transport(GO:1902267) positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.3 | GO:2001014 | regulation of skeletal muscle cell differentiation(GO:2001014) |
| 0.1 | 0.4 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.2 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.1 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.2 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.1 | 0.4 | GO:0042769 | regulation of histone ubiquitination(GO:0033182) DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.6 | GO:0042632 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.4 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.0 | 0.1 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.3 | GO:0061056 | sclerotome development(GO:0061056) |
| 0.0 | 0.1 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.8 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 1.2 | GO:0042773 | ATP synthesis coupled electron transport(GO:0042773) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.2 | GO:0043649 | dicarboxylic acid catabolic process(GO:0043649) |
| 0.0 | 0.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.2 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.0 | 0.3 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.5 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.3 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.2 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.0 | GO:0010084 | specification of organ axis polarity(GO:0010084) |
| 0.0 | 0.3 | GO:0007257 | activation of JUN kinase activity(GO:0007257) positive regulation of JUN kinase activity(GO:0043507) |
| 0.0 | 0.3 | GO:0071427 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.3 | GO:0042738 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.3 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.4 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.0 | 0.7 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 0.3 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.1 | GO:0001845 | phagolysosome assembly(GO:0001845) |
| 0.0 | 0.1 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.1 | GO:0061055 | myotome development(GO:0061055) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0034456 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.2 | 0.8 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.1 | 4.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.6 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.1 | 0.5 | GO:0071005 | U2-type precatalytic spliceosome(GO:0071005) U2-type catalytic step 1 spliceosome(GO:0071006) catalytic step 1 spliceosome(GO:0071012) |
| 0.1 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.4 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.3 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.6 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.1 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0071256 | translocon complex(GO:0071256) |
| 0.0 | 0.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.3 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.2 | 0.6 | GO:0044620 | ACP phosphopantetheine attachment site binding involved in fatty acid biosynthetic process(GO:0000036) ACP phosphopantetheine attachment site binding(GO:0044620) prosthetic group binding(GO:0051192) |
| 0.2 | 4.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.1 | 0.3 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.3 | GO:0102345 | 3-hydroxyacyl-CoA dehydratase activity(GO:0018812) 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.1 | 0.4 | GO:1990404 | protein ADP-ribosylase activity(GO:1990404) |
| 0.1 | 0.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 1.2 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.5 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.3 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.3 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.2 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
| 0.0 | 0.4 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.3 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.2 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.2 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.3 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.3 | GO:0004438 | phosphatidylinositol-3-phosphatase activity(GO:0004438) |
| 0.0 | 0.3 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.6 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.1 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.6 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.4 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.2 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.3 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.1 | 0.2 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.3 | REACTOME ACTIVATION OF NMDA RECEPTOR UPON GLUTAMATE BINDING AND POSTSYNAPTIC EVENTS | Genes involved in Activation of NMDA receptor upon glutamate binding and postsynaptic events |
| 0.0 | 0.6 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.3 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |