Project
DANIO-CODE
Navigation
Downloads
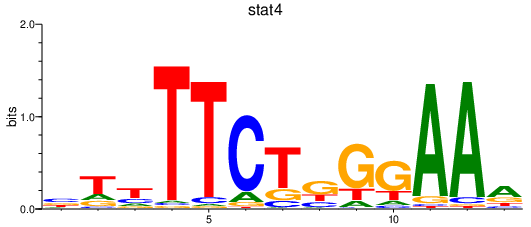
Results for stat4
Z-value: 0.54
Transcription factors associated with stat4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
stat4
|
ENSDARG00000028731 | signal transducer and activator of transcription 4 |
Activity profile of stat4 motif
Sorted Z-values of stat4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of stat4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_23327219 | 0.66 |
ENSDART00000142721
|
dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) |
| chr17_+_24421150 | 0.52 |
ENSDART00000168926
|
mdh1ab
|
malate dehydrogenase 1Ab, NAD (soluble) |
| chr25_+_6059393 | 0.45 |
ENSDART00000154658
|
oaz2a
|
ornithine decarboxylase antizyme 2a |
| chr1_-_11591843 | 0.45 |
ENSDART00000003825
|
cplx2l
|
complexin 2, like |
| chr7_+_39173765 | 0.45 |
ENSDART00000173748
|
tnnt3b
|
troponin T type 3b (skeletal, fast) |
| chr15_+_25554119 | 0.44 |
ENSDART00000162482
|
zgc:152863
|
zgc:152863 |
| chr12_+_23742199 | 0.41 |
|
|
|
| chr15_-_9655500 | 0.41 |
ENSDART00000171214
ENSDART00000171911 |
nars2
|
asparaginyl-tRNA synthetase 2, mitochondrial (putative) |
| chr5_+_29193876 | 0.40 |
ENSDART00000045410
|
thy1
|
Thy-1 cell surface antigen |
| chr4_-_19027117 | 0.40 |
ENSDART00000166160
|
si:dkey-31f5.11
|
si:dkey-31f5.11 |
| chr24_+_8596066 | 0.39 |
ENSDART00000113493
|
tmem14ca
|
transmembrane protein 14Ca |
| chr7_-_28425307 | 0.38 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr9_-_43178744 | 0.38 |
|
|
|
| chr5_-_29818204 | 0.37 |
ENSDART00000098285
|
atf5a
|
activating transcription factor 5a |
| chr5_+_49724855 | 0.37 |
ENSDART00000167163
|
slf1
|
SMC5-SMC6 complex localization factor 1 |
| chr16_-_36090863 | 0.36 |
|
|
|
| chr18_+_17736565 | 0.36 |
ENSDART00000090608
|
rspry1
|
ring finger and SPRY domain containing 1 |
| chr9_+_4188263 | 0.35 |
ENSDART00000164543
|
kalrna
|
kalirin RhoGEF kinase a |
| chr8_-_38126693 | 0.35 |
ENSDART00000112331
|
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr6_+_41506220 | 0.34 |
ENSDART00000136538
|
cish
|
cytokine inducible SH2-containing protein |
| chr6_-_14821305 | 0.34 |
|
|
|
| chr13_+_13550656 | 0.34 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr1_+_41523935 | 0.34 |
ENSDART00000110860
|
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr13_+_1595986 | 0.34 |
|
|
|
| chr19_-_3880783 | 0.33 |
ENSDART00000169639
|
ENSDARG00000101034
|
ENSDARG00000101034 |
| chr22_+_26991665 | 0.33 |
ENSDART00000112826
|
tmem186
|
transmembrane protein 186 |
| chr13_+_2774422 | 0.33 |
ENSDART00000162362
|
wu:fj16a03
|
wu:fj16a03 |
| chr23_+_10534968 | 0.32 |
ENSDART00000140557
|
tns2a
|
tensin 2a |
| chr16_-_31763183 | 0.32 |
ENSDART00000169109
|
rbp5
|
retinol binding protein 1a, cellular |
| chr17_-_12343975 | 0.31 |
ENSDART00000149551
|
emilin1b
|
elastin microfibril interfacer 1b |
| chr5_-_28090077 | 0.31 |
|
|
|
| chr5_-_33265038 | 0.31 |
ENSDART00000132634
|
BX005445.1
|
ENSDARG00000095064 |
| chr22_-_13325409 | 0.31 |
ENSDART00000154095
|
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr1_-_11591684 | 0.29 |
ENSDART00000003825
|
cplx2l
|
complexin 2, like |
| chr19_+_712139 | 0.28 |
ENSDART00000093281
|
fhod3a
|
formin homology 2 domain containing 3a |
| chr16_-_21981065 | 0.28 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr8_+_39964882 | 0.28 |
ENSDART00000134452
|
ggt5a
|
gamma-glutamyltransferase 5a |
| chr2_+_3115593 | 0.28 |
ENSDART00000160715
|
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr6_+_41506504 | 0.27 |
ENSDART00000084861
|
cish
|
cytokine inducible SH2-containing protein |
| chr25_+_3551924 | 0.26 |
ENSDART00000154348
|
prnprs3
|
prion protein, related sequence 3 |
| chr21_-_32027717 | 0.26 |
ENSDART00000131651
|
ENSDARG00000073961
|
ENSDARG00000073961 |
| chr21_-_25528699 | 0.25 |
ENSDART00000144917
|
ENSDARG00000089836
|
ENSDARG00000089836 |
| chr23_-_20402258 | 0.25 |
ENSDART00000136204
|
CR749762.2
|
ENSDARG00000093223 |
| chr22_+_26543146 | 0.25 |
|
|
|
| chr19_+_7255491 | 0.25 |
ENSDART00000151220
|
hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr1_-_23767216 | 0.24 |
ENSDART00000140197
|
sh3d19
|
SH3 domain containing 19 |
| chr19_+_7254917 | 0.24 |
ENSDART00000123934
|
hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr5_-_56953587 | 0.23 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr24_-_16895179 | 0.23 |
ENSDART00000048823
|
c8g
|
complement component 8, gamma polypeptide |
| chr23_-_24556810 | 0.23 |
ENSDART00000109248
|
spen
|
spen family transcriptional repressor |
| chr12_-_37124892 | 0.23 |
ENSDART00000146142
|
pmp22b
|
peripheral myelin protein 22b |
| chr22_-_11463487 | 0.22 |
ENSDART00000002691
|
tspan7b
|
tetraspanin 7b |
| chr16_-_22669752 | 0.22 |
ENSDART00000134239
|
si:dkey-238m4.3
|
si:dkey-238m4.3 |
| chr12_+_26344770 | 0.22 |
ENSDART00000006908
|
itgb3b
|
integrin beta 3b |
| chr9_-_16125551 | 0.22 |
ENSDART00000099479
|
crfb4
|
cytokine receptor family member b4 |
| chr8_+_47908640 | 0.22 |
ENSDART00000140266
|
mfn2
|
mitofusin 2 |
| chr9_+_38692733 | 0.22 |
ENSDART00000087229
|
lmln
|
leishmanolysin-like (metallopeptidase M8 family) |
| chr14_+_34440799 | 0.22 |
ENSDART00000173371
|
sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr7_-_24604255 | 0.22 |
ENSDART00000173920
|
adad2
|
adenosine deaminase domain containing 2 |
| chr22_-_11049321 | 0.22 |
ENSDART00000006927
ENSDART00000170509 |
use1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr19_-_808425 | 0.22 |
ENSDART00000082454
|
glmp
|
glycosylated lysosomal membrane protein |
| chr6_+_7165326 | 0.22 |
ENSDART00000065500
|
abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr22_+_26736632 | 0.21 |
|
|
|
| chr5_+_36168475 | 0.21 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr20_+_23643050 | 0.21 |
ENSDART00000168690
|
palld
|
palladin, cytoskeletal associated protein |
| chr19_+_40792612 | 0.21 |
ENSDART00000017917
|
vps50
|
VPS50 EARP/GARPII complex subunit |
| chr2_-_10300329 | 0.21 |
ENSDART00000138081
|
bcl6ab
|
B-cell CLL/lymphoma 6a, genome duplicate b |
| chr6_+_15123660 | 0.21 |
ENSDART00000155951
|
si:ch73-23l24.1
|
si:ch73-23l24.1 |
| chr24_+_24777962 | 0.20 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr12_+_20578266 | 0.20 |
ENSDART00000016099
|
ENSDARG00000043818
|
ENSDARG00000043818 |
| chr6_+_41506446 | 0.20 |
ENSDART00000140108
|
cish
|
cytokine inducible SH2-containing protein |
| chr25_-_27399421 | 0.19 |
ENSDART00000156906
|
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr7_-_26246473 | 0.19 |
ENSDART00000140694
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr3_-_55586479 | 0.19 |
|
|
|
| chr7_-_57727148 | 0.19 |
|
|
|
| chr12_+_28774063 | 0.18 |
ENSDART00000166229
|
phkg2
|
phosphorylase kinase, gamma 2 (testis) |
| chr13_-_8968218 | 0.18 |
ENSDART00000144142
ENSDART00000144714 |
HTRA2 (1 of many)
|
HtrA serine peptidase 2 |
| chr19_-_808376 | 0.18 |
ENSDART00000140119
|
glmp
|
glycosylated lysosomal membrane protein |
| chr2_-_50244008 | 0.18 |
ENSDART00000029412
|
hecw1b
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1b |
| chr14_-_32291093 | 0.18 |
ENSDART00000167282
ENSDART00000052938 |
atp11c
|
ATPase, Class VI, type 11C |
| chr6_+_41506350 | 0.18 |
ENSDART00000136538
|
cish
|
cytokine inducible SH2-containing protein |
| chr20_+_19879839 | 0.17 |
ENSDART00000132576
|
BX510657.1
|
ENSDARG00000093899 |
| chr2_-_42185736 | 0.17 |
ENSDART00000056460
|
gbp1
|
guanylate binding protein 1 |
| chr5_-_7978801 | 0.17 |
ENSDART00000159477
ENSDART00000171886 ENSDART00000157423 |
lifra
|
leukemia inhibitory factor receptor alpha a |
| chr1_+_11541423 | 0.17 |
ENSDART00000170760
|
tmod1
|
tropomodulin 1 |
| chr20_-_37733350 | 0.17 |
|
|
|
| chr7_-_26246669 | 0.17 |
ENSDART00000140694
|
eif4a1a
|
eukaryotic translation initiation factor 4A1A |
| chr19_-_3547196 | 0.17 |
ENSDART00000166107
|
hivep1
|
human immunodeficiency virus type I enhancer binding protein 1 |
| chr3_-_61144538 | 0.16 |
ENSDART00000155932
|
si:dkey-111k8.2
|
si:dkey-111k8.2 |
| chr3_+_24145201 | 0.16 |
ENSDART00000169765
|
pdgfbb
|
platelet-derived growth factor beta polypeptide b |
| chr23_+_26995808 | 0.16 |
|
|
|
| chr22_+_1445302 | 0.16 |
ENSDART00000164685
|
si:dkeyp-53d3.5
|
si:dkeyp-53d3.5 |
| chr5_-_29818325 | 0.16 |
ENSDART00000098285
|
atf5a
|
activating transcription factor 5a |
| chr16_-_10180957 | 0.15 |
ENSDART00000104037
|
ptpn2a
|
protein tyrosine phosphatase, non-receptor type 2, a |
| chr10_-_7813660 | 0.15 |
ENSDART00000158559
|
si:ch73-340n8.4
|
si:ch73-340n8.4 |
| chr17_-_32913432 | 0.15 |
ENSDART00000077476
|
prox1a
|
prospero homeobox 1a |
| chr23_+_10535195 | 0.15 |
ENSDART00000140557
|
tns2a
|
tensin 2a |
| chr15_+_27454643 | 0.15 |
ENSDART00000164887
ENSDART00000018603 |
tbx4
|
T-box 4 |
| chr13_-_31516399 | 0.15 |
ENSDART00000172375
ENSDART00000125987 ENSDART00000143903 ENSDART00000158719 |
six4a
|
SIX homeobox 4a |
| chr19_-_36009321 | 0.15 |
ENSDART00000136811
|
col8a2
|
collagen, type VIII, alpha 2 |
| chr9_-_16125697 | 0.15 |
ENSDART00000099479
|
crfb4
|
cytokine receptor family member b4 |
| chr2_+_42155813 | 0.15 |
ENSDART00000143562
|
gbp2
|
guanylate binding protein 2 |
| chr17_-_43526955 | 0.14 |
ENSDART00000133665
|
mrpl35
|
mitochondrial ribosomal protein L35 |
| chr12_-_26317480 | 0.14 |
ENSDART00000105636
|
myoz1b
|
myozenin 1b |
| chr21_-_5852663 | 0.14 |
ENSDART00000130521
|
CACFD1
|
calcium channel flower domain containing 1 |
| chr12_-_926596 | 0.14 |
ENSDART00000088351
|
SPAG9 (1 of many)
|
sperm associated antigen 9 |
| chr6_-_40700033 | 0.13 |
ENSDART00000113196
|
si:ch211-157b11.14
|
si:ch211-157b11.14 |
| chr19_+_20209561 | 0.13 |
ENSDART00000168833
|
CR382300.1
|
ENSDARG00000098798 |
| chr20_-_23327126 | 0.13 |
ENSDART00000153308
|
dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) |
| chr17_-_8016214 | 0.13 |
ENSDART00000148520
|
syne1b
|
spectrin repeat containing, nuclear envelope 1b |
| chr2_-_16780900 | 0.13 |
ENSDART00000144801
|
atr
|
ATR serine/threonine kinase |
| chr10_-_28632851 | 0.13 |
ENSDART00000137964
|
bbx
|
bobby sox homolog (Drosophila) |
| chr19_-_23259063 | 0.12 |
ENSDART00000052503
|
nudcd1
|
NudC domain containing 1 |
| chr7_-_24778185 | 0.12 |
ENSDART00000135179
|
arl2
|
ADP-ribosylation factor-like 2 |
| chr14_+_34440699 | 0.12 |
ENSDART00000130469
|
sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr2_+_3115541 | 0.12 |
ENSDART00000160715
|
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr19_-_3880722 | 0.12 |
ENSDART00000169639
|
ENSDARG00000101034
|
ENSDARG00000101034 |
| chr6_-_24292584 | 0.12 |
ENSDART00000163965
|
brdt
|
bromodomain, testis-specific |
| chr10_+_16111842 | 0.12 |
ENSDART00000141654
|
megf10
|
multiple EGF-like-domains 10 |
| chr12_+_22536231 | 0.12 |
ENSDART00000163755
|
si:dkey-219e21.4
|
si:dkey-219e21.4 |
| chr5_-_56953716 | 0.12 |
ENSDART00000167892
|
zgc:193711
|
zgc:193711 |
| chr12_+_36415304 | 0.12 |
ENSDART00000164192
ENSDART00000166309 ENSDART00000175409 |
map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr5_-_29480173 | 0.12 |
ENSDART00000140049
|
bco2a
|
beta-carotene oxygenase 2a |
| chr25_-_35713361 | 0.12 |
ENSDART00000137354
|
dus2
|
dihydrouridine synthase 2 |
| chr4_+_346481 | 0.12 |
ENSDART00000169261
|
tmem181
|
transmembrane protein 181 |
| chr8_+_28433869 | 0.12 |
ENSDART00000062716
|
slc52a3
|
solute carrier family 52 (riboflavin transporter), member 3 |
| chr16_-_39209426 | 0.12 |
|
|
|
| chr5_+_37378340 | 0.11 |
ENSDART00000012050
|
apoa1a
|
apolipoprotein A-Ia |
| chr22_+_26737422 | 0.11 |
|
|
|
| chr20_+_19093849 | 0.11 |
ENSDART00000025509
|
tdh
|
L-threonine dehydrogenase |
| chr2_+_4562038 | 0.11 |
ENSDART00000159525
|
bambib
|
BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) b |
| chr7_-_26226628 | 0.11 |
ENSDART00000058910
|
sox19b
|
SRY (sex determining region Y)-box 19b |
| chr2_+_50126676 | 0.11 |
ENSDART00000145483
|
rpl37
|
ribosomal protein L37 |
| chr21_+_19797559 | 0.11 |
ENSDART00000147555
|
ccdc80l2
|
coiled-coil domain containing 80 like 2 |
| chr21_-_40759739 | 0.11 |
ENSDART00000075808
|
apbb3
|
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr9_+_45193290 | 0.11 |
ENSDART00000176175
|
retsatl
|
retinol saturase (all-trans-retinol 13,14-reductase) like |
| chr21_-_27301938 | 0.10 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr24_+_37596873 | 0.10 |
ENSDART00000109221
ENSDART00000165125 |
wdr90
|
WD repeat domain 90 |
| chr9_+_3414293 | 0.10 |
|
|
|
| chr13_-_42410700 | 0.10 |
ENSDART00000134533
|
btaf1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated |
| chr17_-_32913391 | 0.10 |
ENSDART00000077476
|
prox1a
|
prospero homeobox 1a |
| chr20_-_31035185 | 0.10 |
ENSDART00000147359
|
wtap
|
WT1 associated protein |
| chr18_+_17736311 | 0.10 |
ENSDART00000090608
|
rspry1
|
ring finger and SPRY domain containing 1 |
| chr7_-_26653672 | 0.10 |
ENSDART00000124363
|
alx4a
|
ALX homeobox 4a |
| chr7_-_72500748 | 0.10 |
ENSDART00000160523
|
CU929444.1
|
ENSDARG00000099109 |
| chr21_+_19797435 | 0.10 |
ENSDART00000147555
|
ccdc80l2
|
coiled-coil domain containing 80 like 2 |
| chr22_-_13325475 | 0.10 |
ENSDART00000154095
|
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr8_-_5355293 | 0.10 |
ENSDART00000175272
|
CABZ01051142.1
|
ENSDARG00000106687 |
| chr14_+_28167213 | 0.09 |
ENSDART00000157306
ENSDART00000166375 ENSDART00000167439 ENSDART00000017075 |
xiap
|
X-linked inhibitor of apoptosis |
| chr19_-_27750037 | 0.09 |
ENSDART00000052359
|
gtf2h4
|
general transcription factor IIH, polypeptide 4 |
| chr5_-_7134405 | 0.09 |
ENSDART00000177935
|
bmpr1ba
|
bone morphogenetic protein receptor, type IBa |
| chr20_-_43878454 | 0.09 |
ENSDART00000100610
ENSDART00000100608 ENSDART00000148809 |
matn3a
|
matrilin 3a |
| chr13_-_44492897 | 0.09 |
|
|
|
| chr15_-_12484651 | 0.08 |
ENSDART00000162258
|
BX901957.1
|
ENSDARG00000098363 |
| chr5_-_29818351 | 0.08 |
ENSDART00000098285
|
atf5a
|
activating transcription factor 5a |
| chr5_-_68641376 | 0.08 |
|
|
|
| chr1_+_40758474 | 0.08 |
ENSDART00000139521
|
si:ch211-89o9.4
|
si:ch211-89o9.4 |
| chr21_+_25740782 | 0.08 |
ENSDART00000021620
|
cldnd
|
claudin d |
| chr10_+_21789200 | 0.07 |
|
|
|
| chr7_+_45691346 | 0.07 |
ENSDART00000165691
|
pop4
|
POP4 homolog, ribonuclease P/MRP subunit |
| chr7_+_34216359 | 0.07 |
ENSDART00000173854
|
cln6a
|
CLN6, transmembrane ER protein a |
| chr25_+_32419062 | 0.07 |
|
|
|
| chr5_-_37359835 | 0.07 |
ENSDART00000141059
|
mpzl2b
|
myelin protein zero-like 2b |
| chr6_-_3837294 | 0.07 |
ENSDART00000059212
|
unc50
|
unc-50 homolog (C. elegans) |
| chr2_+_3115688 | 0.07 |
ENSDART00000160715
|
pik3r3a
|
phosphoinositide-3-kinase, regulatory subunit 3a (gamma) |
| chr23_+_24556960 | 0.07 |
|
|
|
| chr9_-_37940101 | 0.07 |
ENSDART00000087663
|
sema5ba
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Ba |
| chr23_-_25853364 | 0.07 |
ENSDART00000110670
|
ENSDARG00000078574
|
ENSDARG00000078574 |
| chr6_-_30702168 | 0.07 |
ENSDART00000065211
|
pars2
|
prolyl-tRNA synthetase 2, mitochondrial (putative) |
| chr24_+_24778026 | 0.07 |
ENSDART00000065288
|
pcyt1ba
|
phosphate cytidylyltransferase 1, choline, beta a |
| chr23_-_25853331 | 0.06 |
ENSDART00000110670
|
ENSDARG00000078574
|
ENSDARG00000078574 |
| chr23_+_23093658 | 0.06 |
ENSDART00000146415
ENSDART00000146463 |
samd11
|
sterile alpha motif domain containing 11 |
| chr19_+_28699884 | 0.06 |
ENSDART00000103862
|
mrpl36
|
mitochondrial ribosomal protein L36 |
| chr2_-_9946148 | 0.06 |
ENSDART00000175460
|
dvl3a
|
dishevelled segment polarity protein 3a |
| chr4_+_21485372 | 0.06 |
ENSDART00000041861
|
syt1a
|
synaptotagmin Ia |
| chr23_-_43916621 | 0.06 |
ENSDART00000015777
|
abce1
|
ATP-binding cassette, sub-family E (OABP), member 1 |
| chr13_-_49430733 | 0.06 |
|
|
|
| chr16_+_44711538 | 0.06 |
ENSDART00000011570
|
zgc:101716
|
zgc:101716 |
| chr22_-_28695519 | 0.06 |
ENSDART00000005883
ENSDART00000007791 |
jagn1b
|
jagunal homolog 1b |
| chr25_+_31979756 | 0.06 |
ENSDART00000012600
|
galk2
|
galactokinase 2 |
| chr6_-_3837266 | 0.06 |
ENSDART00000059212
|
unc50
|
unc-50 homolog (C. elegans) |
| chr16_-_31763281 | 0.06 |
ENSDART00000169109
|
rbp5
|
retinol binding protein 1a, cellular |
| chr3_-_31747486 | 0.06 |
ENSDART00000076189
|
zgc:171779
|
zgc:171779 |
| chr8_-_12171880 | 0.05 |
ENSDART00000132824
|
dab2ipa
|
DAB2 interacting protein a |
| chr10_+_16111920 | 0.05 |
ENSDART00000141654
|
megf10
|
multiple EGF-like-domains 10 |
| chr23_+_43916520 | 0.05 |
ENSDART00000149266
ENSDART00000149503 |
anapc10
|
anaphase promoting complex subunit 10 |
| chr10_+_29883772 | 0.05 |
ENSDART00000100032
|
hyou1
|
hypoxia up-regulated 1 |
| chr9_+_2001849 | 0.05 |
ENSDART00000082329
|
evx2
|
even-skipped homeobox 2 |
| chr6_-_24293008 | 0.05 |
ENSDART00000168355
|
brdt
|
bromodomain, testis-specific |
| chr2_-_42185811 | 0.05 |
ENSDART00000140788
|
gbp1
|
guanylate binding protein 1 |
| chr10_+_8443197 | 0.05 |
ENSDART00000158584
|
hvcn1
|
hydrogen voltage-gated channel 1 |
| chr21_-_42872362 | 0.05 |
ENSDART00000171089
ENSDART00000160998 |
stk10
|
serine/threonine kinase 10 |
| chr24_-_5904097 | 0.05 |
ENSDART00000007373
ENSDART00000135124 |
acbd5a
|
acyl-CoA binding domain containing 5a |
| chr17_+_25659010 | 0.05 |
ENSDART00000029703
|
kcnh1a
|
potassium voltage-gated channel, subfamily H (eag-related), member 1a |
| chr17_+_12613335 | 0.05 |
ENSDART00000112469
|
ralgapa1
|
Ral GTPase activating protein, alpha subunit 1 (catalytic) |
| chr10_-_7813594 | 0.05 |
ENSDART00000158559
|
si:ch73-340n8.4
|
si:ch73-340n8.4 |
| chr15_-_9345785 | 0.05 |
ENSDART00000159676
|
trappc4
|
trafficking protein particle complex 4 |
| chr22_-_4715399 | 0.04 |
ENSDART00000122341
ENSDART00000161345 |
zfr2
|
zinc finger RNA binding protein 2 |
| chr22_+_1445227 | 0.04 |
ENSDART00000164685
|
si:dkeyp-53d3.5
|
si:dkeyp-53d3.5 |
| chr6_+_7165300 | 0.04 |
ENSDART00000065500
|
abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 0.4 | GO:0070445 | peripheral nervous system myelin maintenance(GO:0032287) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.1 | 0.3 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.1 | 0.5 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.1 | 0.2 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.1 | 0.3 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.1 | 0.2 | GO:2000650 | negative regulation of sodium ion transmembrane transporter activity(GO:2000650) |
| 0.1 | 0.2 | GO:0044321 | leptin-mediated signaling pathway(GO:0033210) prolactin signaling pathway(GO:0038161) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.8 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.4 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0003347 | epicardial cell to mesenchymal cell transition(GO:0003347) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.1 | GO:0042214 | carotene metabolic process(GO:0016119) carotene catabolic process(GO:0016121) terpene metabolic process(GO:0042214) terpene catabolic process(GO:0046247) |
| 0.0 | 0.1 | GO:0045940 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) steroid esterification(GO:0034433) sterol esterification(GO:0034434) cholesterol esterification(GO:0034435) reverse cholesterol transport(GO:0043691) positive regulation of steroid metabolic process(GO:0045940) |
| 0.0 | 0.1 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.0 | 0.3 | GO:0006956 | complement activation(GO:0006956) |
| 0.0 | 0.5 | GO:0043551 | regulation of lipid kinase activity(GO:0043550) regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.7 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.0 | 0.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.3 | GO:1900449 | regulation of glutamate receptor signaling pathway(GO:1900449) |
| 0.0 | 0.5 | GO:0006595 | polyamine metabolic process(GO:0006595) |
| 0.0 | 0.2 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.2 | GO:0097324 | melanocyte migration(GO:0097324) |
| 0.0 | 0.5 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.4 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.0 | 0.1 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.4 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.3 | GO:0021884 | forebrain neuron development(GO:0021884) |
| 0.0 | 0.3 | GO:0098754 | detoxification(GO:0098754) |
| 0.0 | 0.1 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.4 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.2 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.0 | 0.1 | GO:0002446 | neutrophil mediated immunity(GO:0002446) defense response to fungus(GO:0050832) |
| 0.0 | 0.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.3 | GO:0002183 | cytoplasmic translational initiation(GO:0002183) |
| 0.0 | 0.1 | GO:0060579 | ventral spinal cord interneuron fate commitment(GO:0060579) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.7 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.9 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.0 | 0.1 | GO:0042627 | chylomicron(GO:0042627) |
| 0.0 | 0.9 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.3 | GO:0036374 | peptidyltransferase activity(GO:0000048) glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.5 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.3 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.4 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 0.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.4 | GO:0031013 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0010436 | beta-carotene 15,15'-monooxygenase activity(GO:0003834) carotenoid dioxygenase activity(GO:0010436) |
| 0.0 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0051373 | telethonin binding(GO:0031433) FATZ binding(GO:0051373) |
| 0.0 | 0.7 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.9 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.5 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.5 | GO:0016876 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0071814 | lipoprotein particle binding(GO:0071813) protein-lipid complex binding(GO:0071814) |
| 0.0 | 0.1 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.1 | GO:0033204 | ribonuclease P RNA binding(GO:0033204) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.6 | GO:0005178 | integrin binding(GO:0005178) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 1.0 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |