Project
DANIO-CODE
Navigation
Downloads
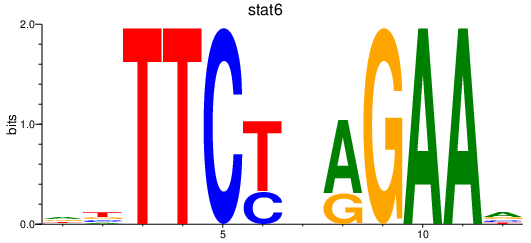
Results for stat6
Z-value: 1.07
Transcription factors associated with stat6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
stat6
|
ENSDARG00000015902 | signal transducer and activator of transcription 6, interleukin-4 induced |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| stat6 | dr10_dc_chr23_-_27196543_27196618 | 0.88 | 7.8e-06 | Click! |
Activity profile of stat6 motif
Sorted Z-values of stat6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of stat6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_16577737 | 4.70 |
ENSDART00000139038
|
CU467861.1
|
ENSDARG00000092337 |
| chr6_-_28231995 | 3.46 |
ENSDART00000131126
|
bcl6a
|
B-cell CLL/lymphoma 6a (zinc finger protein 51) |
| chr19_-_5781358 | 2.78 |
ENSDART00000082050
|
zgc:174904
|
zgc:174904 |
| chr3_-_48865474 | 2.74 |
ENSDART00000133036
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr25_+_30827761 | 2.68 |
ENSDART00000103395
|
tnnt3a
|
troponin T type 3a (skeletal, fast) |
| chr17_-_10682357 | 2.63 |
ENSDART00000064597
|
lgals3b
|
lectin, galactoside binding soluble 3b |
| chr4_+_4795126 | 2.55 |
ENSDART00000129252
|
slc13a4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr3_-_55956030 | 2.39 |
ENSDART00000158163
|
srl
|
sarcalumenin |
| chr10_-_42032702 | 2.33 |
|
|
|
| chr3_+_43010408 | 2.28 |
ENSDART00000169061
|
CU138533.1
|
ENSDARG00000099842 |
| chr2_+_36634309 | 2.26 |
ENSDART00000169547
ENSDART00000098417 |
pak2a
|
p21 protein (Cdc42/Rac)-activated kinase 2a |
| chr17_+_38619041 | 2.24 |
ENSDART00000145147
|
sptb
|
spectrin, beta, erythrocytic |
| chr6_+_6333766 | 2.14 |
ENSDART00000140827
|
bcl11ab
|
B-cell CLL/lymphoma 11Ab |
| chr22_+_26543146 | 2.11 |
|
|
|
| chr24_-_33377293 | 2.09 |
ENSDART00000124938
|
si:ch1073-406l10.2
|
si:ch1073-406l10.2 |
| chr18_-_49324015 | 2.07 |
ENSDART00000098419
|
sb:cb81
|
sb:cb81 |
| chr7_-_7164950 | 2.00 |
ENSDART00000012637
|
zgc:101810
|
zgc:101810 |
| chr7_+_32097831 | 2.00 |
ENSDART00000169588
|
LGR4
|
leucine rich repeat containing G protein-coupled receptor 4 |
| chr2_-_10300329 | 1.97 |
ENSDART00000138081
|
bcl6ab
|
B-cell CLL/lymphoma 6a, genome duplicate b |
| chr7_-_26165200 | 1.95 |
ENSDART00000123395
|
her8a
|
hairy-related 8a |
| chr16_+_40610589 | 1.95 |
ENSDART00000038294
|
tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr7_-_8128948 | 1.93 |
ENSDART00000057101
|
aep1
|
aerolysin-like protein |
| chr24_+_37518715 | 1.91 |
ENSDART00000138264
|
si:ch211-183d21.1
|
si:ch211-183d21.1 |
| chr19_+_43039606 | 1.88 |
ENSDART00000010104
|
crtap
|
cartilage associated protein |
| chr2_-_10667156 | 1.88 |
ENSDART00000081196
|
scinlb
|
scinderin like b |
| chr5_-_42272455 | 1.86 |
ENSDART00000161586
|
flot2a
|
flotillin 2a |
| chr18_-_14891913 | 1.85 |
ENSDART00000018502
|
mapk12a
|
mitogen-activated protein kinase 12a |
| chr1_+_52365033 | 1.83 |
ENSDART00000133411
|
inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr21_-_5852663 | 1.83 |
ENSDART00000130521
|
CACFD1
|
calcium channel flower domain containing 1 |
| chr11_-_17829889 | 1.80 |
ENSDART00000018948
ENSDART00000172343 |
cishb
|
cytokine inducible SH2-containing protein b |
| chr25_+_182037 | 1.77 |
ENSDART00000156469
|
RPS17
|
ribosomal protein S17 |
| chr21_-_32403403 | 1.77 |
ENSDART00000076974
|
gfpt2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr8_-_38126693 | 1.75 |
ENSDART00000112331
|
adgra2
|
adhesion G protein-coupled receptor A2 |
| chr17_+_45824480 | 1.64 |
|
|
|
| chr5_-_25865875 | 1.64 |
ENSDART00000144035
|
arvcfb
|
armadillo repeat gene deleted in velocardiofacial syndrome b |
| chr12_+_21570808 | 1.62 |
|
|
|
| chr25_-_27399421 | 1.59 |
ENSDART00000156906
|
asb15a
|
ankyrin repeat and SOCS box containing 15a |
| chr25_+_35901081 | 1.58 |
ENSDART00000042271
ENSDART00000158027 |
irx3b
|
iroquois homeobox 3b |
| chr16_-_31959111 | 1.57 |
ENSDART00000135669
|
rbfox1l
|
RNA binding protein, fox-1 homolog (C. elegans) 1-like |
| chr11_+_7139675 | 1.55 |
ENSDART00000155864
|
CU929070.1
|
ENSDARG00000097452 |
| chr19_-_3226831 | 1.54 |
ENSDART00000145710
ENSDART00000110763 ENSDART00000074620 |
stm
|
starmaker |
| chr2_+_26632673 | 1.54 |
ENSDART00000017668
|
ptbp1a
|
polypyrimidine tract binding protein 1a |
| chr18_-_15405161 | 1.53 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr7_-_3900223 | 1.52 |
ENSDART00000019949
|
ndrg2
|
NDRG family member 2 |
| chr18_-_15404998 | 1.51 |
ENSDART00000031752
|
rfx4
|
regulatory factor X, 4 |
| chr17_-_12343975 | 1.47 |
ENSDART00000149551
|
emilin1b
|
elastin microfibril interfacer 1b |
| chr7_-_28425307 | 1.44 |
ENSDART00000148822
|
adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr8_-_42617646 | 1.44 |
ENSDART00000075550
|
kazald3
|
Kazal-type serine peptidase inhibitor domain 3 |
| chr1_-_24227846 | 1.43 |
ENSDART00000148076
|
fbxw7
|
F-box and WD repeat domain containing 7 |
| chr13_+_3533517 | 1.42 |
ENSDART00000018737
|
qkib
|
QKI, KH domain containing, RNA binding b |
| chr19_+_43763483 | 1.41 |
ENSDART00000136695
|
yrk
|
Yes-related kinase |
| chr8_-_14054145 | 1.41 |
ENSDART00000137857
|
si:ch211-229n2.7
|
si:ch211-229n2.7 |
| chr14_-_32404076 | 1.38 |
ENSDART00000075617
|
sox3
|
SRY (sex determining region Y)-box 3 |
| chr9_-_34141609 | 1.38 |
|
|
|
| chr10_+_13251463 | 1.37 |
ENSDART00000000887
|
rassf6
|
Ras association (RalGDS/AF-6) domain family 6 |
| chr13_-_1980818 | 1.37 |
ENSDART00000110814
|
fam83b
|
family with sequence similarity 83, member B |
| chr13_+_13550656 | 1.37 |
ENSDART00000057825
|
cfd
|
complement factor D (adipsin) |
| chr6_-_27901354 | 1.37 |
ENSDART00000155116
|
im:7152348
|
im:7152348 |
| chr20_+_30894667 | 1.36 |
ENSDART00000145066
|
nhsl1b
|
NHS-like 1b |
| chr1_-_11591843 | 1.34 |
ENSDART00000003825
|
cplx2l
|
complexin 2, like |
| chr2_+_4562038 | 1.34 |
ENSDART00000159525
|
bambib
|
BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) b |
| chr2_+_4366169 | 1.33 |
ENSDART00000157903
|
gata6
|
GATA binding protein 6 |
| chr5_+_52393600 | 1.33 |
|
|
|
| chr9_-_1949904 | 1.32 |
ENSDART00000082355
|
hoxd4a
|
homeobox D4a |
| chr10_+_32160464 | 1.31 |
ENSDART00000099880
|
wnt11r
|
wingless-type MMTV integration site family, member 11, related |
| chr23_+_23305483 | 1.29 |
ENSDART00000126479
|
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr23_+_2597825 | 1.28 |
ENSDART00000159039
|
ENSDARG00000089986
|
ENSDARG00000089986 |
| chr7_+_30516734 | 1.28 |
ENSDART00000174000
|
apba2b
|
amyloid beta (A4) precursor protein-binding, family A, member 2b |
| chr1_-_54319943 | 1.28 |
ENSDART00000152666
|
slc25a23a
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23a |
| chr21_-_28883441 | 1.27 |
ENSDART00000132884
|
cxxc5a
|
CXXC finger protein 5a |
| chr17_-_22532811 | 1.27 |
ENSDART00000079401
|
si:ch211-125o16.4
|
si:ch211-125o16.4 |
| chr18_+_7607055 | 1.26 |
ENSDART00000140784
|
CR318588.4
|
ENSDARG00000095556 |
| chr23_-_20402258 | 1.25 |
ENSDART00000136204
|
CR749762.2
|
ENSDARG00000093223 |
| chr24_-_40968409 | 1.20 |
ENSDART00000169315
ENSDART00000171543 |
smyhc1
|
slow myosin heavy chain 1 |
| chr12_+_31501522 | 1.20 |
ENSDART00000144422
|
cpn1
|
carboxypeptidase N, polypeptide 1 |
| chr1_+_529491 | 1.19 |
ENSDART00000102421
|
mrpl16
|
mitochondrial ribosomal protein L16 |
| chr2_-_31784388 | 1.19 |
ENSDART00000114928
|
and2
|
actinodin2 |
| chr5_-_56984800 | 1.18 |
ENSDART00000074290
|
mia
|
melanoma inhibitory activity |
| chr25_+_4490129 | 1.18 |
|
|
|
| chr19_+_44185325 | 1.17 |
ENSDART00000063870
|
rpl11
|
ribosomal protein L11 |
| chr19_+_8562864 | 1.15 |
ENSDART00000050217
|
efna1a
|
ephrin-A1a |
| chr23_+_32409339 | 1.15 |
ENSDART00000149698
|
slc39a5
|
solute carrier family 39 (zinc transporter), member 5 |
| chr3_-_47328771 | 1.15 |
ENSDART00000020168
|
kctd5a
|
potassium channel tetramerization domain containing 5a |
| chr17_+_32619043 | 1.15 |
ENSDART00000087565
|
eva1a
|
eva-1 homolog A (C. elegans) |
| chr12_-_650506 | 1.14 |
ENSDART00000172651
|
sult2st2
|
sulfotransferase family 2, cytosolic sulfotransferase 2 |
| chr19_+_12995955 | 1.12 |
ENSDART00000132892
|
cthrc1a
|
collagen triple helix repeat containing 1a |
| chr11_-_11925832 | 1.12 |
|
|
|
| chr23_-_5785468 | 1.12 |
ENSDART00000033093
|
lad1
|
ladinin |
| chr23_-_4990023 | 1.12 |
ENSDART00000142699
|
taz
|
tafazzin |
| chr20_-_25726868 | 1.11 |
ENSDART00000126716
|
paics
|
phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase |
| chr14_+_5852295 | 1.11 |
ENSDART00000128638
|
bscl2l
|
Bernardinelli-Seip congenital lipodystrophy 2, like |
| chr2_+_16492315 | 1.11 |
ENSDART00000135783
|
selj
|
selenoprotein J |
| chr23_-_18958008 | 1.11 |
ENSDART00000133419
|
CR847953.1
|
ENSDARG00000057403 |
| chr5_-_25866099 | 1.11 |
ENSDART00000144035
|
arvcfb
|
armadillo repeat gene deleted in velocardiofacial syndrome b |
| chr13_-_11535112 | 1.10 |
ENSDART00000102411
|
dctn1b
|
dynactin 1b |
| chr8_-_17480730 | 1.09 |
ENSDART00000100667
|
skia
|
v-ski avian sarcoma viral oncogene homolog a |
| chr20_+_23643050 | 1.09 |
ENSDART00000168690
|
palld
|
palladin, cytoskeletal associated protein |
| chr17_+_45594103 | 1.08 |
ENSDART00000153922
|
si:ch211-202f3.3
|
si:ch211-202f3.3 |
| chr10_-_5016621 | 1.06 |
ENSDART00000101414
|
hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr15_+_548292 | 1.05 |
ENSDART00000178563
|
ftr86
|
finTRIM family, member 86 |
| chr11_+_15905877 | 1.03 |
ENSDART00000111284
|
agtrap
|
angiotensin II receptor-associated protein |
| chr15_-_40391882 | 1.03 |
|
|
|
| chr5_+_15319430 | 1.03 |
ENSDART00000162003
|
hspb8
|
heat shock protein b8 |
| chr13_+_3533318 | 1.03 |
ENSDART00000018737
|
qkib
|
QKI, KH domain containing, RNA binding b |
| chr14_-_35332747 | 1.02 |
ENSDART00000074710
|
pdgfc
|
platelet derived growth factor c |
| chr13_+_3533426 | 1.01 |
ENSDART00000018737
|
qkib
|
QKI, KH domain containing, RNA binding b |
| chr8_-_1904720 | 1.01 |
ENSDART00000081563
|
si:dkey-178e17.3
|
si:dkey-178e17.3 |
| chr11_-_8158013 | 1.01 |
ENSDART00000024046
|
uox
|
urate oxidase |
| chr9_+_41238450 | 1.01 |
ENSDART00000000693
|
osgepl1
|
O-sialoglycoprotein endopeptidase-like 1 |
| chr8_+_28513670 | 1.00 |
ENSDART00000110291
|
srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chr17_+_10161999 | 1.00 |
ENSDART00000170274
|
foxa1
|
forkhead box A1 |
| chr9_+_219478 | 0.99 |
ENSDART00000164048
ENSDART00000161484 ENSDART00000171623 |
map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr18_+_37238113 | 0.99 |
ENSDART00000088247
|
qpctla
|
glutaminyl-peptide cyclotransferase-like a |
| chr16_+_4288700 | 0.98 |
ENSDART00000159694
|
inpp5b
|
inositol polyphosphate-5-phosphatase B |
| chr5_-_71352034 | 0.98 |
ENSDART00000007827
|
spra
|
sepiapterin reductase a |
| chr6_+_28232386 | 0.97 |
ENSDART00000163057
|
FP101882.1
|
ENSDARG00000098121 |
| chr20_-_38237357 | 0.96 |
|
|
|
| chr14_-_31128891 | 0.96 |
|
|
|
| chr19_+_15538967 | 0.95 |
ENSDART00000171403
|
lin28a
|
lin-28 homolog A (C. elegans) |
| chr25_-_16890563 | 0.94 |
ENSDART00000073419
|
ccnd2a
|
cyclin D2, a |
| chr15_-_15532980 | 0.94 |
ENSDART00000004220
ENSDART00000131259 |
rab34a
|
RAB34, member RAS oncogene family a |
| chr22_-_13325475 | 0.94 |
ENSDART00000154095
|
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr2_-_42185736 | 0.94 |
ENSDART00000056460
|
gbp1
|
guanylate binding protein 1 |
| chr3_-_62156768 | 0.93 |
ENSDART00000140782
|
proza
|
protein Z, vitamin K-dependent plasma glycoprotein a |
| chr13_+_49527181 | 0.93 |
ENSDART00000176643
|
CABZ01084653.1
|
ENSDARG00000106801 |
| chr20_+_28958586 | 0.93 |
ENSDART00000142678
|
slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr16_+_33189439 | 0.92 |
ENSDART00000101953
|
rhbdl2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr1_-_11591684 | 0.92 |
ENSDART00000003825
|
cplx2l
|
complexin 2, like |
| chr1_-_58606982 | 0.92 |
ENSDART00000158067
|
txndc11
|
thioredoxin domain containing 11 |
| chr11_-_17829827 | 0.91 |
ENSDART00000018948
ENSDART00000172343 |
cishb
|
cytokine inducible SH2-containing protein b |
| chr10_+_404464 | 0.91 |
ENSDART00000145124
ENSDART00000124319 |
dact3a
|
dishevelled-binding antagonist of beta-catenin 3a |
| chr2_-_42185811 | 0.91 |
ENSDART00000140788
|
gbp1
|
guanylate binding protein 1 |
| chr22_-_13325409 | 0.91 |
ENSDART00000154095
|
si:ch211-227m13.1
|
si:ch211-227m13.1 |
| chr20_-_42306410 | 0.91 |
ENSDART00000074959
|
slc35f1
|
solute carrier family 35, member F1 |
| chr23_+_17939650 | 0.91 |
ENSDART00000162822
|
naca
|
nascent polypeptide-associated complex alpha subunit |
| chr24_-_16895179 | 0.91 |
ENSDART00000048823
|
c8g
|
complement component 8, gamma polypeptide |
| chr21_-_27301938 | 0.91 |
ENSDART00000130632
|
hif1al2
|
hypoxia-inducible factor 1, alpha subunit, like 2 |
| chr6_-_51538315 | 0.91 |
|
|
|
| chr8_-_22321957 | 0.90 |
ENSDART00000137687
|
smim1
|
small integral membrane protein 1 |
| chr7_-_57727148 | 0.90 |
|
|
|
| chr15_-_47260896 | 0.89 |
ENSDART00000098711
|
eif3k
|
eukaryotic translation initiation factor 3, subunit K |
| chr5_-_25865936 | 0.89 |
ENSDART00000144035
|
arvcfb
|
armadillo repeat gene deleted in velocardiofacial syndrome b |
| chr1_-_25704525 | 0.88 |
ENSDART00000171614
|
ppa2
|
pyrophosphatase (inorganic) 2 |
| chr18_+_16257606 | 0.88 |
ENSDART00000142584
|
alx1
|
ALX homeobox 1 |
| chr21_+_8249235 | 0.88 |
ENSDART00000129749
|
psmb7
|
proteasome subunit beta 7 |
| chr3_+_25933164 | 0.87 |
ENSDART00000143697
|
si:dkeyp-69e1.8
|
si:dkeyp-69e1.8 |
| chr14_+_30390575 | 0.87 |
ENSDART00000087884
|
ccdc85b
|
coiled-coil domain containing 85B |
| chr21_-_9313788 | 0.87 |
|
|
|
| chr15_+_31042182 | 0.87 |
ENSDART00000153754
|
BX000438.2
|
ENSDARG00000098011 |
| chr10_+_10252074 | 0.87 |
ENSDART00000144214
|
sh2d3ca
|
SH2 domain containing 3Ca |
| chr23_+_10534968 | 0.86 |
ENSDART00000140557
|
tns2a
|
tensin 2a |
| chr20_+_19175518 | 0.86 |
|
|
|
| chr9_-_33330725 | 0.86 |
|
|
|
| chr16_-_21981065 | 0.86 |
ENSDART00000078858
|
si:ch73-86n18.1
|
si:ch73-86n18.1 |
| chr1_+_14451504 | 0.86 |
ENSDART00000111475
|
pcdh7a
|
protocadherin 7a |
| chr11_+_6284324 | 0.85 |
ENSDART00000002459
|
ctns
|
cystinosin, lysosomal cystine transporter |
| chr5_-_43334777 | 0.85 |
ENSDART00000142271
|
ENSDARG00000053091
|
ENSDARG00000053091 |
| chr20_-_7303735 | 0.85 |
ENSDART00000100060
|
dsc2l
|
desmocollin 2 like |
| chr3_-_32727588 | 0.85 |
ENSDART00000158916
|
si:dkey-16l2.20
|
si:dkey-16l2.20 |
| chr21_-_4842528 | 0.84 |
ENSDART00000097796
|
rnf165a
|
ring finger protein 165a |
| chr8_+_47909655 | 0.84 |
|
|
|
| chr13_+_28574593 | 0.84 |
ENSDART00000126845
|
ldb1a
|
LIM domain binding 1a |
| chr2_-_50244008 | 0.83 |
ENSDART00000029412
|
hecw1b
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1b |
| chr12_-_9479063 | 0.83 |
ENSDART00000169727
|
si:ch211-207i20.3
|
si:ch211-207i20.3 |
| chr9_-_10561062 | 0.83 |
ENSDART00000175269
|
thsd7ba
|
thrombospondin, type I, domain containing 7Ba |
| chr7_+_48047139 | 0.83 |
ENSDART00000052122
|
crabp1b
|
cellular retinoic acid binding protein 1b |
| chr25_-_14573221 | 0.83 |
|
|
|
| chr15_+_42626315 | 0.82 |
|
|
|
| chr21_+_26695270 | 0.81 |
|
|
|
| chr19_-_2486568 | 0.81 |
ENSDART00000012791
|
sp8a
|
sp8 transcription factor a |
| chr4_-_671973 | 0.80 |
ENSDART00000164735
|
rtcb
|
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
| chr5_+_69036145 | 0.80 |
ENSDART00000174403
|
fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr20_+_52653111 | 0.79 |
ENSDART00000057980
|
tsta3
|
tissue specific transplantation antigen P35B |
| chr23_-_38228305 | 0.79 |
ENSDART00000131791
|
pfdn4
|
prefoldin subunit 4 |
| chr8_+_8899112 | 0.79 |
ENSDART00000145970
|
si:dkey-83k24.5
|
si:dkey-83k24.5 |
| chr18_-_38235232 | 0.79 |
ENSDART00000114224
|
si:dkey-10o6.2
|
si:dkey-10o6.2 |
| chr4_-_18447913 | 0.79 |
ENSDART00000141671
|
socs2
|
suppressor of cytokine signaling 2 |
| chr8_+_39762186 | 0.79 |
ENSDART00000143413
|
si:ch211-170d8.2
|
si:ch211-170d8.2 |
| chr1_+_11448327 | 0.79 |
ENSDART00000126020
|
spink4
|
serine peptidase inhibitor, Kazal type 4 |
| chr19_+_712139 | 0.79 |
ENSDART00000093281
|
fhod3a
|
formin homology 2 domain containing 3a |
| chr25_-_13057808 | 0.78 |
ENSDART00000172571
|
smpd3
|
sphingomyelin phosphodiesterase 3, neutral |
| chr5_+_44407763 | 0.77 |
ENSDART00000010786
|
dmrt2a
|
doublesex and mab-3 related transcription factor 2a |
| chr14_+_7626822 | 0.77 |
ENSDART00000109941
|
cxxc5b
|
CXXC finger protein 5b |
| chr4_-_13903253 | 0.77 |
ENSDART00000032805
|
gxylt1b
|
glucoside xylosyltransferase 1b |
| chr15_+_23615727 | 0.77 |
ENSDART00000152720
|
MARK4
|
microtubule affinity regulating kinase 4 |
| chr20_-_7303804 | 0.76 |
ENSDART00000100060
|
dsc2l
|
desmocollin 2 like |
| chr8_+_584353 | 0.76 |
ENSDART00000048498
|
CABZ01049925.1
|
ENSDARG00000079790 |
| chr5_+_26804344 | 0.76 |
ENSDART00000121886
ENSDART00000005025 |
hdr
|
hematopoietic death receptor |
| chr6_-_8075384 | 0.76 |
ENSDART00000129674
|
slc44a2
|
solute carrier family 44 (choline transporter), member 2 |
| chr24_+_42884 | 0.76 |
ENSDART00000122785
|
zgc:152808
|
zgc:152808 |
| chr2_-_43997672 | 0.75 |
ENSDART00000005449
|
zeb1a
|
zinc finger E-box binding homeobox 1a |
| chr3_-_30554400 | 0.75 |
ENSDART00000151097
|
si:ch211-51c14.1
|
si:ch211-51c14.1 |
| chr4_-_8005840 | 0.74 |
ENSDART00000036153
|
ccdc3a
|
coiled-coil domain containing 3a |
| chr24_+_28441089 | 0.74 |
|
|
|
| chr21_-_32403459 | 0.73 |
ENSDART00000076974
|
gfpt2
|
glutamine-fructose-6-phosphate transaminase 2 |
| chr25_-_34602827 | 0.73 |
ENSDART00000124121
|
hist1h4l
|
histone 1, H4, like |
| chr3_-_48865399 | 0.73 |
ENSDART00000028610
|
elavl3
|
ELAV like neuron-specific RNA binding protein 3 |
| chr16_+_40558786 | 0.72 |
ENSDART00000126129
|
nagpa
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase |
| chr1_+_1657493 | 0.72 |
ENSDART00000103850
|
atp1a1a.3
|
ATPase, Na+/K+ transporting, alpha 1a polypeptide, tandem duplicate 3 |
| chr3_+_39514598 | 0.70 |
ENSDART00000156038
|
epn2
|
epsin 2 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.8 | 3.0 | GO:0021910 | smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021910) |
| 0.5 | 1.4 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.5 | 1.4 | GO:0031642 | negative regulation of myelination(GO:0031642) negative regulation of neurological system process(GO:0031645) |
| 0.5 | 1.4 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.4 | 1.7 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.4 | 1.6 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.4 | 1.9 | GO:1904086 | regulation of epiboly involved in gastrulation with mouth forming second(GO:1904086) |
| 0.3 | 1.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.3 | 1.3 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.3 | 1.3 | GO:1902306 | negative regulation of sodium ion transmembrane transport(GO:1902306) |
| 0.3 | 1.6 | GO:0035777 | pronephric distal tubule development(GO:0035777) |
| 0.3 | 1.5 | GO:0007343 | egg activation(GO:0007343) |
| 0.3 | 0.9 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.3 | 4.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.3 | 1.6 | GO:0045299 | otolith mineralization(GO:0045299) |
| 0.2 | 0.7 | GO:0070587 | regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.2 | 1.0 | GO:0071871 | response to monoamine(GO:0071867) response to catecholamine(GO:0071869) response to epinephrine(GO:0071871) |
| 0.2 | 0.9 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.2 | 1.7 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.2 | 2.5 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.2 | 0.6 | GO:0070640 | vitamin D3 metabolic process(GO:0070640) |
| 0.2 | 2.1 | GO:0050994 | regulation of lipid catabolic process(GO:0050994) |
| 0.2 | 0.5 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.2 | 1.0 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.2 | 1.3 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.2 | 1.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.2 | 0.5 | GO:0044320 | leptin-mediated signaling pathway(GO:0033210) prolactin signaling pathway(GO:0038161) cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.2 | 0.9 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.2 | 3.2 | GO:0006942 | regulation of striated muscle contraction(GO:0006942) |
| 0.1 | 0.6 | GO:0035332 | positive regulation of hippo signaling(GO:0035332) |
| 0.1 | 0.9 | GO:0070293 | renal absorption(GO:0070293) modified amino acid transport(GO:0072337) |
| 0.1 | 1.1 | GO:0032048 | cardiolipin metabolic process(GO:0032048) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.1 | 1.4 | GO:0006956 | complement activation(GO:0006956) |
| 0.1 | 0.4 | GO:0032212 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.1 | 1.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.1 | 0.5 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) |
| 0.1 | 1.6 | GO:0060975 | cardioblast migration(GO:0003260) cardioblast migration to the midline involved in heart field formation(GO:0060975) |
| 0.1 | 0.9 | GO:0048823 | nucleate erythrocyte development(GO:0048823) |
| 0.1 | 0.8 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 1.0 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.1 | 1.9 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.1 | 0.4 | GO:1902514 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.1 | 1.4 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.1 | 4.6 | GO:0030834 | regulation of actin filament depolymerization(GO:0030834) negative regulation of actin filament depolymerization(GO:0030835) actin filament capping(GO:0051693) |
| 0.1 | 0.3 | GO:0045905 | positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.7 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.1 | 2.4 | GO:1903845 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.1 | 0.4 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.1 | 1.0 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.1 | 0.5 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 0.5 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 1.0 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 1.4 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 0.3 | GO:0050886 | sleep(GO:0030431) endocrine process(GO:0050886) |
| 0.1 | 2.3 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 2.1 | GO:0001558 | regulation of cell growth(GO:0001558) |
| 0.1 | 1.1 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 0.9 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.1 | 0.5 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 2.1 | GO:0050773 | regulation of dendrite development(GO:0050773) |
| 0.1 | 0.5 | GO:1900027 | regulation of ruffle assembly(GO:1900027) |
| 0.1 | 0.8 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.2 | GO:0035776 | pronephric proximal tubule development(GO:0035776) |
| 0.1 | 1.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.1 | 0.2 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.1 | 0.2 | GO:0048659 | smooth muscle cell proliferation(GO:0048659) |
| 0.1 | 1.2 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.1 | 0.9 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.8 | GO:0000394 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 0.3 | GO:0070782 | phosphatidylserine exposure on apoptotic cell surface(GO:0070782) |
| 0.1 | 1.1 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.1 | 1.3 | GO:0071391 | cellular response to estrogen stimulus(GO:0071391) |
| 0.1 | 0.6 | GO:0006959 | humoral immune response(GO:0006959) |
| 0.1 | 3.9 | GO:0001816 | cytokine production(GO:0001816) |
| 0.1 | 1.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.1 | 0.8 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.5 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.1 | 0.2 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 1.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.1 | 0.7 | GO:0014812 | muscle cell migration(GO:0014812) |
| 0.1 | 1.0 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.6 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.9 | GO:0071456 | cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.3 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 0.7 | GO:0036376 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.0 | 3.2 | GO:0050770 | regulation of axonogenesis(GO:0050770) |
| 0.0 | 3.6 | GO:0050768 | negative regulation of neurogenesis(GO:0050768) |
| 0.0 | 1.4 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 0.9 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.0 | 2.2 | GO:0008285 | negative regulation of cell proliferation(GO:0008285) |
| 0.0 | 0.9 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 1.4 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 2.6 | GO:0006814 | sodium ion transport(GO:0006814) |
| 0.0 | 0.8 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 2.0 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.6 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.0 | 0.8 | GO:0007548 | sex differentiation(GO:0007548) |
| 0.0 | 0.5 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.0 | 0.7 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.4 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 1.2 | GO:0051260 | protein homooligomerization(GO:0051260) |
| 0.0 | 0.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 1.3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.8 | GO:0015696 | ammonium transport(GO:0015696) |
| 0.0 | 0.5 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.0 | 1.3 | GO:0034620 | cellular response to unfolded protein(GO:0034620) |
| 0.0 | 0.4 | GO:0044243 | collagen catabolic process(GO:0030574) multicellular organism catabolic process(GO:0044243) |
| 0.0 | 0.1 | GO:0032355 | response to estradiol(GO:0032355) |
| 0.0 | 3.5 | GO:0050767 | regulation of neurogenesis(GO:0050767) |
| 0.0 | 0.1 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.2 | GO:0046512 | diol biosynthetic process(GO:0034312) sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 3.9 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.7 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 0.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.9 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) positive regulation of proteasomal protein catabolic process(GO:1901800) |
| 0.0 | 1.2 | GO:0006821 | chloride transport(GO:0006821) |
| 0.0 | 0.1 | GO:0030326 | embryonic limb morphogenesis(GO:0030326) |
| 0.0 | 0.3 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.9 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.6 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 0.6 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.7 | GO:0030100 | regulation of endocytosis(GO:0030100) |
| 0.0 | 0.9 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 1.5 | GO:0002040 | sprouting angiogenesis(GO:0002040) |
| 0.0 | 1.5 | GO:0043062 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.8 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 1.2 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.4 | GO:0007041 | lysosomal transport(GO:0007041) |
| 0.0 | 1.2 | GO:0051216 | cartilage development(GO:0051216) |
| 0.0 | 0.4 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.4 | 2.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.4 | 1.9 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.3 | 0.9 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 0.3 | 1.3 | GO:0043034 | costamere(GO:0043034) |
| 0.2 | 1.9 | GO:0046930 | pore complex(GO:0046930) |
| 0.2 | 1.0 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.2 | 1.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.2 | 0.5 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.2 | 2.3 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 3.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.0 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.8 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 2.0 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.3 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.1 | 0.5 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.1 | 0.6 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) phosphatidylinositol 3-kinase complex, class I(GO:0097651) |
| 0.1 | 1.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 1.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.4 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 1.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.5 | GO:0071256 | translocon complex(GO:0071256) |
| 0.1 | 1.5 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.8 | GO:0036379 | striated muscle thin filament(GO:0005865) myofilament(GO:0036379) |
| 0.1 | 0.4 | GO:0005921 | gap junction(GO:0005921) |
| 0.1 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.0 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.1 | 0.4 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) clathrin-coated endocytic vesicle membrane(GO:0030669) |
| 0.1 | 0.4 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 2.2 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.9 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 1.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 1.6 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 6.0 | GO:0005912 | adherens junction(GO:0005912) anchoring junction(GO:0070161) |
| 0.0 | 1.3 | GO:0043197 | dendritic spine(GO:0043197) neuron spine(GO:0044309) |
| 0.0 | 0.4 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.3 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 2.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 1.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.4 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.2 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 1.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.3 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 14.4 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 0.4 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 0.2 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 0.5 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 1.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 2.6 | GO:0000151 | ubiquitin ligase complex(GO:0000151) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0019863 | IgE binding(GO:0019863) immunoglobulin binding(GO:0019865) |
| 0.8 | 2.5 | GO:0004360 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity(GO:0004360) L-glutamine aminotransferase activity(GO:0070548) |
| 0.4 | 3.2 | GO:0030172 | troponin C binding(GO:0030172) troponin I binding(GO:0031013) |
| 0.4 | 1.9 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.3 | 0.9 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.3 | 0.9 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.3 | 1.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.2 | 2.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 1.8 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.2 | 1.1 | GO:0004639 | phosphoribosylaminoimidazolesuccinocarboxamide synthase activity(GO:0004639) |
| 0.2 | 0.9 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.2 | 1.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 0.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 0.5 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.2 | 0.8 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.2 | 0.8 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.1 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 1.0 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.1 | 0.5 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 1.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.1 | 1.0 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 1.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 0.7 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.1 | 0.8 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.1 | 0.5 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 2.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.2 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.1 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.1 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.3 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 1.9 | GO:0005518 | collagen binding(GO:0005518) |
| 0.1 | 0.5 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 1.3 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 4.8 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.1 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 2.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.1 | 1.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.1 | 0.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 1.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.9 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 1.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.8 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 1.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.1 | 0.3 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.1 | 0.3 | GO:0034595 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) |
| 0.1 | 0.3 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.1 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.1 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 1.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 2.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.0 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.5 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 1.2 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.3 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 1.3 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 2.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 1.1 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 2.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 8.3 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.4 | GO:0019201 | nucleotide kinase activity(GO:0019201) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.5 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 1.1 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.5 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 1.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 2.5 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 0.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 1.0 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.0 | 0.7 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 2.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.4 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.8 | GO:0046332 | SMAD binding(GO:0046332) |
| 0.0 | 0.6 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.3 | GO:0009931 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) calcium-dependent protein serine/threonine kinase activity(GO:0009931) calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.9 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 5.9 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 0.6 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.4 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 2.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.9 | GO:0051082 | unfolded protein binding(GO:0051082) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.4 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 1.6 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.3 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.7 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.8 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 2.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.2 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.6 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.1 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.0 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.3 | 2.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.2 | 1.0 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 2.5 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 1.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 0.9 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.1 | 0.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 1.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 2.3 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 0.5 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 0.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 2.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.6 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.8 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.7 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.8 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.5 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.6 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.5 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 1.8 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.3 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.4 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.4 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.2 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |