Project
DANIO-CODE
Navigation
Downloads
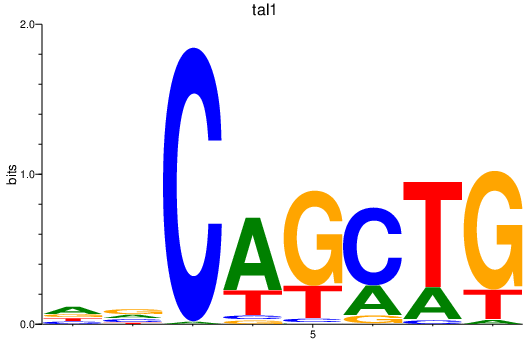
Results for tal1
Z-value: 1.09
Transcription factors associated with tal1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tal1
|
ENSDARG00000019930 | T-cell acute lymphocytic leukemia 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tal1 | dr10_dc_chr22_+_16509286_16509322 | 0.85 | 3.1e-05 | Click! |
Activity profile of tal1 motif
Sorted Z-values of tal1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of tal1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_9115745 | 3.15 |
ENSDART00000123652
|
hmgn6
|
high mobility group nucleosome binding domain 6 |
| chr8_-_985673 | 2.89 |
ENSDART00000170737
|
smyd1b
|
SET and MYND domain containing 1b |
| chr2_-_16896965 | 2.75 |
ENSDART00000022549
|
atp1b3a
|
ATPase, Na+/K+ transporting, beta 3a polypeptide |
| chr25_+_30699938 | 2.67 |
ENSDART00000036275
|
tnni2a.4
|
troponin I type 2a (skeletal, fast), tandem duplicate 4 |
| chr25_-_35819345 | 2.46 |
ENSDART00000064400
|
si:ch211-113a14.24
|
si:ch211-113a14.24 |
| chr11_-_22211478 | 2.45 |
ENSDART00000065996
|
tmem183a
|
transmembrane protein 183A |
| chr3_-_58170499 | 2.32 |
ENSDART00000074272
|
im:6904045
|
im:6904045 |
| chr25_+_31457309 | 2.31 |
ENSDART00000022325
|
parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr7_+_23778570 | 2.22 |
ENSDART00000121837
|
efs
|
embryonal Fyn-associated substrate |
| chr16_-_42040346 | 2.21 |
ENSDART00000169313
|
pycard
|
PYD and CARD domain containing |
| chr16_+_24057260 | 2.19 |
ENSDART00000132742
ENSDART00000145330 |
apoc1
|
apolipoprotein C-I |
| chr14_-_36523075 | 2.12 |
ENSDART00000109293
|
rnf130
|
ring finger protein 130 |
| chr1_+_27174549 | 2.12 |
ENSDART00000102337
|
dnajb14
|
DnaJ (Hsp40) homolog, subfamily B, member 14 |
| chr23_+_565406 | 2.10 |
ENSDART00000055134
|
ogfr
|
opioid growth factor receptor |
| chr24_-_26165778 | 2.07 |
ENSDART00000080113
|
apodb
|
apolipoprotein Db |
| chr9_+_308260 | 2.06 |
ENSDART00000163474
|
stac3
|
SH3 and cysteine rich domain 3 |
| chr20_+_2443682 | 1.98 |
ENSDART00000131642
|
akap7
|
A kinase (PRKA) anchor protein 7 |
| chr17_-_9917832 | 1.96 |
ENSDART00000161574
ENSDART00000166081 |
zgc:109986
|
zgc:109986 |
| chr21_+_27346176 | 1.92 |
ENSDART00000005682
|
actn3a
|
actinin alpha 3a |
| chr17_+_135165 | 1.90 |
ENSDART00000166339
|
klhdc2
|
kelch domain containing 2 |
| chr5_+_40722565 | 1.89 |
ENSDART00000097546
|
arid3c
|
AT rich interactive domain 3C (BRIGHT-like) |
| chr2_-_42784609 | 1.84 |
|
|
|
| chr22_+_33177432 | 1.77 |
ENSDART00000126885
|
dag1
|
dystroglycan 1 |
| chr9_+_51693805 | 1.75 |
ENSDART00000132896
|
fap
|
fibroblast activation protein, alpha |
| chr1_+_51925783 | 1.68 |
ENSDART00000167514
|
abca1a
|
ATP-binding cassette, sub-family A (ABC1), member 1A |
| chr3_-_1463509 | 1.67 |
ENSDART00000053892
|
ENSDARG00000074266
|
ENSDARG00000074266 |
| chr22_-_26333957 | 1.65 |
ENSDART00000130493
|
capn2b
|
calpain 2, (m/II) large subunit b |
| chr25_+_182037 | 1.65 |
ENSDART00000156469
|
RPS17
|
ribosomal protein S17 |
| chr16_+_22950567 | 1.64 |
ENSDART00000143957
|
flad1
|
flavin adenine dinucleotide synthetase 1 |
| chr17_+_53068731 | 1.61 |
ENSDART00000156774
|
dph6
|
diphthamine biosynthesis 6 |
| chr8_-_23172938 | 1.56 |
ENSDART00000141090
|
si:ch211-196c10.15
|
si:ch211-196c10.15 |
| chr25_-_34648956 | 1.56 |
ENSDART00000154851
|
zgc:153405
|
zgc:153405 |
| chr22_+_26543146 | 1.56 |
|
|
|
| chr17_+_53224434 | 1.53 |
ENSDART00000166517
|
asb2b
|
ankyrin repeat and SOCS box containing 2b |
| chr23_+_36822250 | 1.53 |
|
|
|
| chr12_-_25825072 | 1.53 |
|
|
|
| KN150475v1_-_1819 | 1.52 |
|
|
|
| chr3_+_54327353 | 1.52 |
ENSDART00000127487
|
eif3g
|
eukaryotic translation initiation factor 3, subunit G |
| chr7_+_6517248 | 1.52 |
ENSDART00000102681
|
pnp5a
|
purine nucleoside phosphorylase 5a |
| chr25_+_34545289 | 1.51 |
ENSDART00000126625
|
si:dkey-108k21.10
|
si:dkey-108k21.10 |
| chr22_-_120677 | 1.48 |
|
|
|
| KN149858v1_-_12281 | 1.47 |
ENSDART00000157456
|
CABZ01056637.1
|
ENSDARG00000100203 |
| chr4_+_8637319 | 1.46 |
|
|
|
| chr9_+_16437408 | 1.43 |
ENSDART00000006787
|
epha3
|
eph receptor A3 |
| chr16_+_55167489 | 1.41 |
ENSDART00000149795
|
nr0b2a
|
nuclear receptor subfamily 0, group B, member 2a |
| chr14_-_8975187 | 1.36 |
ENSDART00000054693
|
asb12b
|
ankyrin repeat and SOCS box-containing 12b |
| chr3_-_29846530 | 1.36 |
ENSDART00000077111
|
hsd17b14
|
hydroxysteroid (17-beta) dehydrogenase 14 |
| chr16_+_54751779 | 1.32 |
|
|
|
| chr6_-_51573878 | 1.31 |
ENSDART00000166911
|
rbl1
|
retinoblastoma-like 1 (p107) |
| chr22_+_34738389 | 1.30 |
ENSDART00000154372
|
hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr7_-_6334472 | 1.30 |
ENSDART00000081359
|
zgc:110425
|
zgc:110425 |
| chr3_-_25886553 | 1.29 |
ENSDART00000030890
|
hmox1a
|
heme oxygenase 1a |
| chr24_-_26138298 | 1.27 |
ENSDART00000128618
|
and1
|
actinodin1 |
| chr19_+_31817286 | 1.27 |
ENSDART00000078459
|
tmem55a
|
transmembrane protein 55A |
| chr16_-_41553835 | 1.24 |
ENSDART00000029492
|
cmtm7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr15_+_29343644 | 1.24 |
ENSDART00000170537
ENSDART00000128973 |
rap1gap2a
|
RAP1 GTPase activating protein 2a |
| chr14_-_46648563 | 1.22 |
|
|
|
| chr1_-_23765358 | 1.22 |
|
|
|
| chr10_+_4987494 | 1.21 |
ENSDART00000121959
|
si:ch73-234b20.5
|
si:ch73-234b20.5 |
| chr4_-_5294280 | 1.20 |
ENSDART00000178921
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr19_-_24971633 | 1.19 |
ENSDART00000162801
|
polr3gla
|
polymerase (RNA) III (DNA directed) polypeptide G like a |
| chr16_+_5625301 | 1.17 |
|
|
|
| chr16_+_3090170 | 1.17 |
ENSDART00000110395
|
limd1a
|
LIM domains containing 1a |
| chr3_-_6922655 | 1.17 |
|
|
|
| chr14_+_28126921 | 1.17 |
ENSDART00000026846
|
pin4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| KN149932v1_+_27584 | 1.16 |
|
|
|
| chr7_+_32451041 | 1.15 |
ENSDART00000126565
|
si:ch211-150g13.3
|
si:ch211-150g13.3 |
| chr3_+_6150498 | 1.13 |
ENSDART00000175915
|
BX284638.2
|
ENSDARG00000108888 |
| chr14_-_25279835 | 1.13 |
ENSDART00000163669
|
cplx2
|
complexin 2 |
| chr6_+_4712785 | 1.12 |
ENSDART00000151674
|
pcdh9
|
protocadherin 9 |
| chr11_-_80746 | 1.12 |
|
|
|
| chr19_-_22182031 | 1.11 |
ENSDART00000104279
|
znf516
|
zinc finger protein 516 |
| chr21_-_36338514 | 1.10 |
ENSDART00000157826
|
mpp1
|
membrane protein, palmitoylated 1 |
| chr2_+_43736542 | 1.09 |
ENSDART00000148454
|
itgb1b.1
|
integrin, beta 1b.1 |
| chr8_+_29733109 | 1.07 |
ENSDART00000020621
|
mapk4
|
mitogen-activated protein kinase 4 |
| chr15_-_12484651 | 1.06 |
ENSDART00000162258
|
BX901957.1
|
ENSDARG00000098363 |
| chr22_+_223797 | 1.05 |
|
|
|
| chr15_+_28435937 | 1.05 |
ENSDART00000142298
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr19_+_48499602 | 1.04 |
|
|
|
| chr8_+_28513670 | 1.04 |
ENSDART00000110291
|
srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chr2_+_1602501 | 1.03 |
ENSDART00000109668
|
bcl10
|
B-cell CLL/lymphoma 10 |
| chr12_+_16235258 | 1.00 |
ENSDART00000141169
|
ppp1r3cb
|
protein phosphatase 1, regulatory subunit 3Cb |
| chr21_-_4842528 | 0.99 |
ENSDART00000097796
|
rnf165a
|
ring finger protein 165a |
| chr2_+_4366169 | 0.98 |
ENSDART00000157903
|
gata6
|
GATA binding protein 6 |
| chr12_-_36138709 | 0.98 |
ENSDART00000130985
|
st6galnac
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase |
| chr24_+_17125429 | 0.97 |
ENSDART00000017605
|
spag6
|
sperm associated antigen 6 |
| chr2_+_16492315 | 0.96 |
ENSDART00000135783
|
selj
|
selenoprotein J |
| KN150475v1_-_1760 | 0.96 |
|
|
|
| chr11_-_11317608 | 0.96 |
ENSDART00000113311
|
col9a1a
|
collagen, type IX, alpha 1a |
| chr11_-_25803101 | 0.95 |
ENSDART00000088888
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr11_-_25693072 | 0.93 |
ENSDART00000122011
|
kaznb
|
kazrin, periplakin interacting protein b |
| chr7_-_6296880 | 0.93 |
ENSDART00000172954
|
si:ch1073-153i20.4
|
si:ch1073-153i20.4 |
| chr23_+_22729939 | 0.93 |
ENSDART00000009337
|
eno1a
|
enolase 1a, (alpha) |
| chr16_-_33637966 | 0.93 |
ENSDART00000142965
|
BX511161.1
|
ENSDARG00000092272 |
| chr24_-_26254551 | 0.93 |
ENSDART00000112317
|
zgc:194621
|
zgc:194621 |
| chr10_+_16634200 | 0.92 |
ENSDART00000101142
|
chsy3
|
chondroitin sulfate synthase 3 |
| chr12_-_650506 | 0.91 |
ENSDART00000172651
|
sult2st2
|
sulfotransferase family 2, cytosolic sulfotransferase 2 |
| chr8_-_27667972 | 0.91 |
ENSDART00000086946
|
mov10b.1
|
Moloney leukemia virus 10b, tandem duplicate 1 |
| chr6_+_59749159 | 0.90 |
|
|
|
| chr12_-_3270927 | 0.90 |
ENSDART00000175918
|
CABZ01063170.1
|
ENSDARG00000107441 |
| chr1_+_21309690 | 0.90 |
ENSDART00000141317
|
dnah6
|
dynein, axonemal, heavy chain 6 |
| chr18_-_54695 | 0.89 |
|
|
|
| chr5_-_60967429 | 0.88 |
ENSDART00000050914
|
si:dkey-261j4.4
|
si:dkey-261j4.4 |
| chr6_+_6640324 | 0.88 |
ENSDART00000150967
|
si:ch211-85n16.3
|
si:ch211-85n16.3 |
| chr10_-_36864268 | 0.88 |
ENSDART00000165853
|
dhrs13a.1
|
dehydrogenase/reductase (SDR family) member 13a, tandem duplicate 1 |
| chr5_+_71611136 | 0.87 |
ENSDART00000163906
|
CABZ01115396.1
|
ENSDARG00000099805 |
| chr8_+_999710 | 0.87 |
ENSDART00000081432
|
sprb
|
sepiapterin reductase b |
| chr11_+_666006 | 0.86 |
ENSDART00000130839
|
timp4.1
|
TIMP metallopeptidase inhibitor 4, tandem duplicate 1 |
| chr20_+_34417665 | 0.85 |
ENSDART00000153207
|
ivns1abpa
|
influenza virus NS1A binding protein a |
| chr2_-_44402486 | 0.85 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase, Na+/K+ transporting, alpha 2a polypeptide |
| chr16_+_7709183 | 0.85 |
ENSDART00000065514
|
stx12l
|
syntaxin 12, like |
| KN150487v1_+_38777 | 0.84 |
|
|
|
| chr23_-_9990121 | 0.84 |
ENSDART00000127029
|
si:ch211-220i18.4
|
si:ch211-220i18.4 |
| chr24_+_252774 | 0.82 |
ENSDART00000178656
ENSDART00000175867 |
CABZ01080224.1
|
ENSDARG00000107432 |
| chr10_-_8336541 | 0.82 |
ENSDART00000163803
|
plpp1a
|
phospholipid phosphatase 1a |
| chr24_-_32750010 | 0.82 |
ENSDART00000038364
|
ca2
|
carbonic anhydrase II |
| chr25_-_31291943 | 0.81 |
ENSDART00000175104
|
lamb4
|
laminin, beta 4 |
| chr7_-_72127164 | 0.81 |
ENSDART00000176069
|
CABZ01057364.1
|
ENSDARG00000106736 |
| chr18_-_26483645 | 0.80 |
|
|
|
| chr5_+_65267043 | 0.79 |
ENSDART00000161578
|
mymk
|
myomaker, myoblast fusion factor |
| chr19_-_2939818 | 0.78 |
|
|
|
| KN150680v1_+_10854 | 0.77 |
|
|
|
| chr4_+_5240888 | 0.77 |
ENSDART00000150391
|
si:ch211-214j24.14
|
si:ch211-214j24.14 |
| chr5_-_67083987 | 0.77 |
ENSDART00000167301
|
b4galt4
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 4 |
| chr6_+_53349584 | 0.77 |
ENSDART00000167079
|
si:ch211-161c3.5
|
si:ch211-161c3.5 |
| chr5_+_51201924 | 0.76 |
ENSDART00000087467
|
cmya5
|
cardiomyopathy associated 5 |
| chr4_-_16365281 | 0.76 |
ENSDART00000139919
|
lum
|
lumican |
| chr2_+_21267793 | 0.75 |
ENSDART00000099913
|
pla2g4aa
|
phospholipase A2, group IVAa (cytosolic, calcium-dependent) |
| chr15_+_2590598 | 0.75 |
ENSDART00000178505
|
SH2B2
|
SH2B adaptor protein 2 |
| chr12_+_22605268 | 0.74 |
|
|
|
| chr9_-_23936295 | 0.74 |
ENSDART00000165907
|
ndufa10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10 |
| chr4_+_45204456 | 0.74 |
ENSDART00000158744
|
BX927336.3
|
ENSDARG00000102848 |
| KN149874v1_+_3898 | 0.74 |
|
|
|
| chr22_+_5657904 | 0.74 |
ENSDART00000138102
|
dnase1l4.2
|
deoxyribonuclease 1 like 4, tandem duplicate 2 |
| chr14_-_46648119 | 0.74 |
|
|
|
| chr21_+_45325383 | 0.73 |
ENSDART00000029946
|
ube2b
|
ubiquitin-conjugating enzyme E2B (RAD6 homolog) |
| chr18_+_6664159 | 0.72 |
|
|
|
| chr23_+_29981525 | 0.72 |
ENSDART00000035755
|
dvl1a
|
dishevelled segment polarity protein 1a |
| chr24_+_2740747 | 0.72 |
ENSDART00000169394
|
LYRM4
|
LYR motif containing 4 |
| chr23_-_17543731 | 0.72 |
ENSDART00000002398
|
trim101
|
tripartite motif containing 101 |
| chr13_+_51769487 | 0.71 |
ENSDART00000169928
ENSDART00000160698 |
klhdc3
|
kelch domain containing 3 |
| chr1_+_44414936 | 0.71 |
|
|
|
| chr20_+_6165025 | 0.70 |
ENSDART00000077072
|
CABZ01062780.1
|
ENSDARG00000037190 |
| chr14_-_1081316 | 0.70 |
|
|
|
| chr25_+_30472954 | 0.70 |
|
|
|
| chr25_-_35037680 | 0.69 |
ENSDART00000097651
|
si:ch211-87j1.4
|
si:ch211-87j1.4 |
| chr1_+_157674 | 0.69 |
ENSDART00000152205
ENSDART00000160843 |
cul4a
|
cullin 4A |
| chr3_+_1440282 | 0.69 |
ENSDART00000158633
|
poldip3
|
polymerase (DNA-directed), delta interacting protein 3 |
| chr16_+_5411816 | 0.68 |
|
|
|
| chr22_+_20695983 | 0.68 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr20_+_23392782 | 0.67 |
|
|
|
| chr8_+_52633069 | 0.67 |
ENSDART00000162953
|
cyr61l2
|
cysteine-rich, angiogenic inducer, 61 like 2 |
| chr18_+_91971 | 0.67 |
|
|
|
| chr3_+_17387551 | 0.66 |
ENSDART00000104549
|
hcrt
|
hypocretin (orexin) neuropeptide precursor |
| chr8_-_20357907 | 0.65 |
|
|
|
| chr20_-_53629996 | 0.65 |
ENSDART00000141826
|
mettl24
|
methyltransferase like 24 |
| chr19_-_44043682 | 0.65 |
ENSDART00000138009
ENSDART00000086138 |
fam83hb
|
family with sequence similarity 83, member Hb |
| chr8_-_11806627 | 0.64 |
ENSDART00000064017
|
rapgef1a
|
Rap guanine nucleotide exchange factor (GEF) 1a |
| chr11_+_30048913 | 0.64 |
|
|
|
| chr10_-_24690263 | 0.64 |
ENSDART00000163102
|
proser1
|
proline and serine rich 1 |
| chr23_-_3815492 | 0.64 |
ENSDART00000131536
|
hmga1a
|
high mobility group AT-hook 1a |
| chr22_-_38271070 | 0.64 |
ENSDART00000165430
ENSDART00000059690 |
hps3
|
Hermansky-Pudlak syndrome 3 |
| chr24_-_36449162 | 0.63 |
ENSDART00000154402
|
si:ch211-40k21.9
|
si:ch211-40k21.9 |
| chr10_-_28638101 | 0.63 |
ENSDART00000177781
|
bbx
|
bobby sox homolog (Drosophila) |
| chr21_-_45341242 | 0.62 |
ENSDART00000075438
|
cdkn2aipnl
|
CDKN2A interacting protein N-terminal like |
| chr23_+_22408855 | 0.62 |
ENSDART00000147696
|
rap1gap
|
RAP1 GTPase activating protein |
| chr16_+_42868905 | 0.62 |
ENSDART00000156601
|
si:ch211-135n15.2
|
si:ch211-135n15.2 |
| chr17_+_29316965 | 0.62 |
|
|
|
| chr1_-_54331157 | 0.62 |
ENSDART00000083572
|
zgc:136864
|
zgc:136864 |
| chr14_+_28126739 | 0.62 |
ENSDART00000026846
|
pin4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr11_-_41512092 | 0.61 |
ENSDART00000173147
|
FP325130.3
|
ENSDARG00000105426 |
| chr3_-_62143464 | 0.61 |
ENSDART00000155853
|
gprc5ba
|
G protein-coupled receptor, class C, group 5, member Ba |
| chr7_+_69232260 | 0.61 |
ENSDART00000073861
|
gabarapb
|
GABA(A) receptor-associated protein b |
| chr11_-_11487856 | 0.60 |
ENSDART00000030103
|
krt94
|
keratin 94 |
| chr25_-_727490 | 0.60 |
|
|
|
| chr9_+_33177527 | 0.60 |
ENSDART00000174533
|
vangl1
|
VANGL planar cell polarity protein 1 |
| chr2_+_289138 | 0.60 |
ENSDART00000157246
|
znf1008
|
zinc finger protein 1008 |
| chr7_-_30451749 | 0.59 |
ENSDART00000052539
|
myo1ea
|
myosin IE, a |
| chr24_-_2740524 | 0.59 |
ENSDART00000171805
|
fars2
|
phenylalanyl-tRNA synthetase 2, mitochondrial |
| chr18_+_54352 | 0.59 |
ENSDART00000097163
|
SLC27A2 (1 of many)
|
solute carrier family 27 member 2 |
| chr14_-_46648350 | 0.58 |
|
|
|
| chr12_+_29648614 | 0.58 |
ENSDART00000161533
|
gfra1b
|
gdnf family receptor alpha 1b |
| chr1_-_10423289 | 0.58 |
|
|
|
| chr7_+_1397973 | 0.58 |
|
|
|
| chr15_-_47269988 | 0.57 |
ENSDART00000151594
|
h3f3b.1
|
H3 histone, family 3B.1 |
| chr20_+_9776103 | 0.57 |
ENSDART00000053834
|
psmc6
|
proteasome 26S subunit, ATPase 6 |
| chr20_-_296201 | 0.57 |
ENSDART00000104806
|
wisp3
|
WNT1 inducible signaling pathway protein 3 |
| chr2_-_44402452 | 0.56 |
ENSDART00000011188
ENSDART00000093298 |
atp1a2a
|
ATPase, Na+/K+ transporting, alpha 2a polypeptide |
| chr17_+_135261 | 0.56 |
ENSDART00000166339
|
klhdc2
|
kelch domain containing 2 |
| KN149895v1_+_73008 | 0.56 |
|
|
|
| chr1_-_518543 | 0.55 |
ENSDART00000138032
|
trmt10c
|
tRNA methyltransferase 10C, mitochondrial RNase P subunit |
| chr7_+_48726955 | 0.55 |
ENSDART00000166329
|
si:ch211-288d18.1
|
si:ch211-288d18.1 |
| chr3_-_1421391 | 0.54 |
ENSDART00000149709
|
smdt1a
|
single-pass membrane protein with aspartate-rich tail 1a |
| chr16_+_330953 | 0.54 |
ENSDART00000102977
|
nrsn1
|
neurensin 1 |
| chr14_-_29565869 | 0.54 |
ENSDART00000172349
|
sorbs2b
|
sorbin and SH3 domain containing 2b |
| chr2_-_42510909 | 0.54 |
ENSDART00000144274
|
si:dkey-7l6.3
|
si:dkey-7l6.3 |
| chr2_-_45386850 | 0.53 |
|
|
|
| chr23_-_5080531 | 0.52 |
|
|
|
| chr3_+_39617522 | 0.52 |
|
|
|
| chr20_+_25326042 | 0.52 |
ENSDART00000130494
|
moxd1
|
monooxygenase, DBH-like 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0032369 | negative regulation of lipid transport(GO:0032369) |
| 0.5 | 1.6 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.5 | 2.1 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0014808) sarcoplasmic reticulum calcium ion transport(GO:0070296) calcium ion transport from endoplasmic reticulum to cytosol(GO:1903514) |
| 0.5 | 1.4 | GO:0010084 | specification of organ axis polarity(GO:0010084) |
| 0.4 | 2.2 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.4 | 1.3 | GO:0007585 | respiratory gaseous exchange(GO:0007585) regulation of respiratory gaseous exchange(GO:0043576) |
| 0.4 | 1.7 | GO:0032374 | regulation of sterol transport(GO:0032371) regulation of cholesterol transport(GO:0032374) |
| 0.4 | 1.6 | GO:0006747 | FAD biosynthetic process(GO:0006747) FAD metabolic process(GO:0046443) flavin adenine dinucleotide metabolic process(GO:0072387) flavin adenine dinucleotide biosynthetic process(GO:0072388) |
| 0.4 | 2.7 | GO:0048769 | sarcomerogenesis(GO:0048769) |
| 0.4 | 1.4 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.3 | 0.9 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.3 | 0.8 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.3 | 1.6 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.2 | 0.9 | GO:0032889 | regulation of vacuole fusion, non-autophagic(GO:0032889) |
| 0.2 | 0.7 | GO:0001659 | temperature homeostasis(GO:0001659) sleep(GO:0030431) |
| 0.2 | 1.3 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.2 | 0.6 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.2 | 1.8 | GO:0033292 | T-tubule organization(GO:0033292) |
| 0.2 | 0.8 | GO:0042698 | ovarian follicle development(GO:0001541) ovulation cycle process(GO:0022602) ovulation cycle(GO:0042698) inflammatory response to wounding(GO:0090594) |
| 0.2 | 2.8 | GO:0071436 | cellular potassium ion homeostasis(GO:0030007) sodium ion export from cell(GO:0036376) sodium ion export(GO:0071436) |
| 0.2 | 0.5 | GO:0042423 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.2 | 0.5 | GO:0021610 | glossopharyngeal nerve development(GO:0021563) facial nerve morphogenesis(GO:0021610) |
| 0.2 | 1.2 | GO:2000637 | negative regulation of hippo signaling(GO:0035331) positive regulation of posttranscriptional gene silencing(GO:0060148) positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.2 | 0.9 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.1 | 2.3 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.1 | 0.4 | GO:0097250 | mitochondrial respiratory chain supercomplex assembly(GO:0097250) |
| 0.1 | 1.0 | GO:0001773 | myeloid dendritic cell activation(GO:0001773) myeloid dendritic cell differentiation(GO:0043011) dendritic cell differentiation(GO:0097028) |
| 0.1 | 2.7 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.7 | GO:0090178 | establishment of planar polarity of embryonic epithelium(GO:0042249) establishment of planar polarity involved in neural tube closure(GO:0090177) regulation of establishment of planar polarity involved in neural tube closure(GO:0090178) planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 1.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.1 | 0.9 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.1 | 2.9 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 1.6 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.1 | 0.9 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.8 | GO:0014743 | regulation of muscle hypertrophy(GO:0014743) muscle hypertrophy(GO:0014896) |
| 0.1 | 0.9 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.3 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.1 | 0.5 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 2.1 | GO:0007568 | aging(GO:0007568) |
| 0.1 | 2.2 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.1 | 0.6 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.9 | GO:0051923 | sulfation(GO:0051923) |
| 0.1 | 1.5 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 1.3 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.1 | 0.8 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.1 | 0.3 | GO:0045598 | regulation of fat cell differentiation(GO:0045598) |
| 0.1 | 0.3 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.1 | 1.4 | GO:0001732 | formation of cytoplasmic translation initiation complex(GO:0001732) |
| 0.1 | 1.4 | GO:0071218 | response to misfolded protein(GO:0051788) cellular response to misfolded protein(GO:0071218) |
| 0.1 | 1.1 | GO:0030050 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.1 | 1.1 | GO:0033627 | cell adhesion mediated by integrin(GO:0033627) |
| 0.1 | 0.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 1.0 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) regulation of polysaccharide biosynthetic process(GO:0032885) |
| 0.1 | 1.1 | GO:0046928 | regulation of neurotransmitter secretion(GO:0046928) |
| 0.1 | 0.7 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) |
| 0.1 | 0.7 | GO:2000134 | negative regulation of cell cycle G1/S phase transition(GO:1902807) negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.9 | GO:0060287 | epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 1.0 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.0 | 0.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.4 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.3 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.4 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.7 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.3 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.2 | GO:2000009 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.8 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.5 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 1.0 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 0.2 | GO:0045050 | protein insertion into ER membrane by stop-transfer membrane-anchor sequence(GO:0045050) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0046168 | NADH oxidation(GO:0006116) glycerol-3-phosphate catabolic process(GO:0046168) |
| 0.0 | 1.0 | GO:0045104 | intermediate filament-based process(GO:0045103) intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.2 | GO:0035372 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 1.3 | GO:0048278 | vesicle docking(GO:0048278) |
| 0.0 | 2.0 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.7 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.8 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.8 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.1 | GO:0061001 | regulation of dendritic spine morphogenesis(GO:0061001) |
| 0.0 | 1.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.2 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.5 | 1.6 | GO:0098594 | mucin granule(GO:0098594) |
| 0.3 | 2.8 | GO:0090533 | sodium:potassium-exchanging ATPase complex(GO:0005890) cation-transporting ATPase complex(GO:0090533) ATPase dependent transmembrane transport complex(GO:0098533) ATPase complex(GO:1904949) |
| 0.3 | 1.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.2 | 2.7 | GO:0031430 | M band(GO:0031430) |
| 0.2 | 2.2 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.2 | 0.9 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 1.8 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.2 | 0.6 | GO:0070062 | extracellular exosome(GO:0070062) |
| 0.1 | 2.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.1 | 0.9 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.1 | 2.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.0 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) endoplasmic reticulum subcompartment(GO:0098827) |
| 0.1 | 0.7 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.5 | GO:0032420 | stereocilium(GO:0032420) |
| 0.1 | 0.5 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.1 | 1.1 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.1 | 0.8 | GO:0005605 | basal lamina(GO:0005605) laminin complex(GO:0043256) |
| 0.1 | 0.6 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.1 | 1.4 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) |
| 0.1 | 0.5 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.6 | GO:0042645 | nucleoid(GO:0009295) ribonuclease P complex(GO:0030677) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 1.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 1.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.5 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 1.2 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 1.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.9 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 2.8 | GO:0005924 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) |
| 0.0 | 0.8 | GO:0044853 | caveola(GO:0005901) plasma membrane raft(GO:0044853) |
| 0.0 | 2.1 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.5 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.6 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.7 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 2.6 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.2 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.6 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.5 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.7 | 2.2 | GO:0032090 | Pyrin domain binding(GO:0032090) |
| 0.4 | 1.6 | GO:0003919 | FMN adenylyltransferase activity(GO:0003919) |
| 0.4 | 1.5 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.4 | 1.1 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.3 | 1.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.3 | 1.7 | GO:0090554 | phospholipid-translocating ATPase activity(GO:0004012) phosphatidylcholine-translocating ATPase activity(GO:0090554) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.3 | 2.2 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.3 | 0.8 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.2 | 0.9 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.2 | 1.3 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.2 | 0.9 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.2 | 1.8 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.2 | 0.9 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.2 | 0.9 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.2 | 0.5 | GO:0004088 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.2 | 2.7 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.2 | 6.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 2.8 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.6 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 1.4 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 1.8 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.9 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.4 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 1.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 1.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.5 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 1.4 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.1 | 2.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.1 | 0.4 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 1.0 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 1.6 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.1 | 1.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.7 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.6 | GO:0030295 | kinase activator activity(GO:0019209) protein kinase activator activity(GO:0030295) |
| 0.0 | 0.3 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.8 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 1.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.8 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 1.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.8 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 1.0 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 1.0 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.3 | GO:0016289 | CoA hydrolase activity(GO:0016289) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.9 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 2.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 1.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.5 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.3 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 2.9 | GO:0061659 | ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.3 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.8 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.2 | 1.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.4 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.7 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 1.3 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.5 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.6 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 0.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.9 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.8 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 0.7 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.1 | 1.8 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.1 | 2.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 0.7 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.8 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 1.0 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.5 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.4 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 1.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.5 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 1.0 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |