Project
DANIO-CODE
Navigation
Downloads
Results for tbp
Z-value: 2.45
Transcription factors associated with tbp
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbp
|
ENSDARG00000014994 | TATA box binding protein |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbp | dr10_dc_chr13_-_24265471_24265568 | -0.62 | 1.0e-02 | Click! |
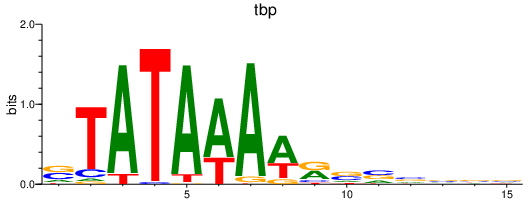
Activity profile of tbp motif
Sorted Z-values of tbp motif
Network of associatons between targets according to the STRING database.
First level regulatory network of tbp
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_45950530 | 10.39 |
ENSDART00000060822
|
afp4
|
antifreeze protein type IV |
| chr2_+_17112049 | 10.11 |
ENSDART00000125647
|
tfa
|
transferrin-a |
| chr5_+_37378340 | 9.16 |
ENSDART00000012050
|
apoa1a
|
apolipoprotein A-Ia |
| chr3_-_32686790 | 8.90 |
ENSDART00000075465
|
mylpfa
|
myosin light chain, phosphorylatable, fast skeletal muscle a |
| chr5_-_31674204 | 8.41 |
ENSDART00000122889
|
myhz1.3
|
myosin, heavy polypeptide 1.3, skeletal muscle |
| chr12_-_17590507 | 8.35 |
ENSDART00000010144
|
pvalb2
|
parvalbumin 2 |
| chr8_-_16577737 | 7.55 |
ENSDART00000139038
|
CU467861.1
|
ENSDARG00000092337 |
| chr8_+_933677 | 6.97 |
ENSDART00000149528
|
fabp1b.1
|
fatty acid binding protein 1b, tandem duplicate 1 |
| chr16_+_24045774 | 6.15 |
ENSDART00000133484
|
apoeb
|
apolipoprotein Eb |
| chr5_+_4033390 | 6.10 |
ENSDART00000149185
|
sat1a.2
|
spermidine/spermine N1-acetyltransferase 1a, duplicate 2 |
| chr5_-_24982964 | 5.90 |
ENSDART00000031665
ENSDART00000161043 |
anxa1a
|
annexin A1a |
| chr2_-_7693224 | 5.75 |
ENSDART00000167019
|
wu:fc46h12
|
wu:fc46h12 |
| chr11_+_7139675 | 5.72 |
ENSDART00000155864
|
CU929070.1
|
ENSDARG00000097452 |
| chr9_-_22434581 | 5.62 |
ENSDART00000114943
|
crygm2d4
|
crystallin, gamma M2d4 |
| KN150173v1_-_16439 | 5.58 |
ENSDART00000162810
|
SNORD60
|
Small nucleolar RNA SNORD60 |
| chr23_-_10242377 | 5.50 |
ENSDART00000129044
|
krt5
|
keratin 5 |
| chr5_+_68410884 | 5.42 |
ENSDART00000153691
|
CU928129.1
|
ENSDARG00000097815 |
| chr14_-_46651359 | 5.41 |
ENSDART00000163316
|
CR383676.1
|
ENSDARG00000099970 |
| chr5_-_70948223 | 5.40 |
ENSDART00000013404
|
ak1
|
adenylate kinase 1 |
| chr8_+_26799298 | 5.31 |
ENSDART00000134987
|
si:ch211-156j16.1
|
si:ch211-156j16.1 |
| chr9_-_45802484 | 5.26 |
ENSDART00000139019
|
agr1
|
anterior gradient 1 |
| chr5_-_36237656 | 5.17 |
ENSDART00000032481
|
ckma
|
creatine kinase, muscle a |
| chr18_-_45750 | 5.12 |
ENSDART00000148821
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr24_-_41448993 | 5.07 |
ENSDART00000041349
|
crygn2
|
crystallin, gamma N2 |
| chr11_+_6446302 | 4.96 |
ENSDART00000140707
ENSDART00000036939 |
gadd45ba
|
growth arrest and DNA-damage-inducible, beta a |
| chr9_-_22402341 | 4.88 |
ENSDART00000110656
|
crygm2d20
|
crystallin, gamma M2d20 |
| chr4_+_828545 | 4.86 |
ENSDART00000067461
|
si:ch211-152c2.3
|
si:ch211-152c2.3 |
| chr9_-_22470612 | 4.79 |
ENSDART00000123763
|
crygm2d3
|
crystallin, gamma M2d3 |
| chr4_-_26333625 | 4.72 |
ENSDART00000170709
|
st8sia1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| KN149795v1_-_24393 | 4.67 |
|
|
|
| chr2_+_24852094 | 4.65 |
ENSDART00000052063
|
rps28
|
ribosomal protein S28 |
| chr6_+_251026 | 4.63 |
ENSDART00000042970
|
atf4a
|
activating transcription factor 4a |
| chr19_-_5429428 | 4.62 |
ENSDART00000066620
ENSDART00000151398 |
krtt1c19e
|
keratin type 1 c19e |
| chr6_-_54172509 | 4.61 |
ENSDART00000149468
|
rps10
|
ribosomal protein S10 |
| chr25_+_34515221 | 4.56 |
ENSDART00000046218
|
flnca
|
filamin C, gamma a (actin binding protein 280) |
| chr10_-_6816534 | 4.56 |
ENSDART00000110735
|
zgc:194281
|
zgc:194281 |
| chr9_-_22371512 | 4.16 |
ENSDART00000101809
|
crygm2d6
|
crystallin, gamma M2d6 |
| chr23_-_21520413 | 4.14 |
ENSDART00000044080
ENSDART00000112929 |
her12
|
hairy-related 12 |
| chr9_-_22507592 | 4.09 |
ENSDART00000132029
|
crygm2d2
|
crystallin, gamma M2d2 |
| chr8_+_6819898 | 4.07 |
ENSDART00000101450
ENSDART00000064163 |
nefmb
|
neurofilament, medium polypeptide b |
| chr3_+_15355472 | 4.06 |
ENSDART00000141714
|
nupr1
|
nuclear protein 1 |
| chr13_-_12513113 | 4.03 |
|
|
|
| chr9_-_22318918 | 3.99 |
ENSDART00000124272
|
crygm2d17
|
crystallin, gamma M2d17 |
| chr24_-_1307905 | 3.98 |
ENSDART00000159212
|
nrp1a
|
neuropilin 1a |
| chr23_+_19887319 | 3.96 |
ENSDART00000139192
|
emd
|
emerin (Emery-Dreifuss muscular dystrophy) |
| chr19_+_17832515 | 3.93 |
ENSDART00000149045
|
klf2b
|
Kruppel-like factor 2b |
| chr24_+_32261533 | 3.91 |
ENSDART00000003745
|
vim
|
vimentin |
| chr9_-_52847086 | 3.81 |
ENSDART00000171721
|
dap1b
|
death associated protein 1b |
| chr9_-_22488422 | 3.76 |
ENSDART00000124600
|
crygm2d21
|
crystallin, gamma M2d21 |
| chr2_+_24881103 | 3.76 |
ENSDART00000144149
|
angptl4
|
angiopoietin-like 4 |
| chr17_-_52020689 | 3.74 |
ENSDART00000019766
|
tgfb3
|
transforming growth factor, beta 3 |
| chr10_-_44180588 | 3.64 |
ENSDART00000145404
|
crybb1
|
crystallin, beta B1 |
| chr15_+_28435937 | 3.63 |
ENSDART00000142298
|
slc43a2a
|
solute carrier family 43 (amino acid system L transporter), member 2a |
| chr22_+_26543146 | 3.63 |
|
|
|
| KN150074v1_-_1031 | 3.63 |
|
|
|
| chr11_-_11591394 | 3.58 |
ENSDART00000142208
|
zgc:110712
|
zgc:110712 |
| chr23_+_27749014 | 3.57 |
ENSDART00000128833
|
rps26
|
ribosomal protein S26 |
| chr9_-_22324699 | 3.57 |
ENSDART00000175417
ENSDART00000101902 |
crygm2d8
|
crystallin, gamma M2d8 |
| chr20_-_54192057 | 3.56 |
ENSDART00000023550
|
hsp90aa1.2
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 2 |
| chr23_+_25378921 | 3.55 |
ENSDART00000143291
|
RPL41
|
ribosomal protein L41 |
| chr5_-_21523520 | 3.54 |
ENSDART00000023306
|
asb12a
|
ankyrin repeat and SOCS box-containing 12a |
| chr15_-_5827067 | 3.54 |
ENSDART00000102459
|
rbp2a
|
retinol binding protein 2a, cellular |
| chr14_+_46419051 | 3.51 |
ENSDART00000112377
|
map1lc3cl
|
microtubule-associated protein 1 light chain 3 gamma, like |
| chr20_-_9992898 | 3.46 |
ENSDART00000080664
|
ACTC1
|
zgc:86709 |
| chr9_-_22306541 | 3.46 |
ENSDART00000101916
|
crygm2d15
|
crystallin, gamma M2d15 |
| chr21_-_28486800 | 3.46 |
ENSDART00000077910
|
epdl2
|
ependymin-like 2 |
| chr22_+_16471319 | 3.42 |
ENSDART00000014330
|
ier5
|
immediate early response 5 |
| chr2_-_38017902 | 3.37 |
ENSDART00000034790
|
pcp4l1
|
Purkinje cell protein 4 like 1 |
| chr6_+_35378839 | 3.37 |
ENSDART00000102483
ENSDART00000133783 |
rgs4
|
regulator of G protein signaling 4 |
| chr9_-_1701626 | 3.36 |
ENSDART00000163482
|
hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chr22_-_10372023 | 3.34 |
|
|
|
| chr19_-_16126342 | 3.29 |
|
|
|
| chr6_-_43094573 | 3.18 |
ENSDART00000084389
|
lrrn1
|
leucine rich repeat neuronal 1 |
| chr9_-_22461196 | 3.05 |
ENSDART00000135190
|
crygm2d7
|
crystallin, gamma M2d7 |
| chr8_-_31359923 | 3.01 |
ENSDART00000019937
|
gadd45ga
|
growth arrest and DNA-damage-inducible, gamma a |
| chr15_+_548292 | 2.95 |
ENSDART00000178563
|
ftr86
|
finTRIM family, member 86 |
| chr3_-_31672763 | 2.95 |
ENSDART00000028270
|
gfap
|
glial fibrillary acidic protein |
| chr1_+_54460288 | 2.90 |
ENSDART00000138070
ENSDART00000150510 |
mb
|
myoglobin |
| chr25_-_13580057 | 2.86 |
ENSDART00000090226
|
znf319b
|
zinc finger protein 319b |
| chr18_-_45892 | 2.81 |
ENSDART00000052641
|
gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr9_-_22347397 | 2.79 |
ENSDART00000146188
|
crygm2d16
|
crystallin, gamma M2d16 |
| chr24_+_21476448 | 2.79 |
ENSDART00000002595
|
rpl21
|
ribosomal protein L21 |
| chr8_+_48624273 | 2.78 |
ENSDART00000121432
|
nppa
|
natriuretic peptide A |
| chr1_+_9999842 | 2.76 |
ENSDART00000152438
|
ENSDARG00000037679
|
ENSDARG00000037679 |
| chr18_+_26102262 | 2.76 |
ENSDART00000164495
|
znf710a
|
zinc finger protein 710a |
| chr8_+_46651079 | 2.72 |
ENSDART00000113803
|
her3
|
hairy-related 3 |
| chr11_-_6960107 | 2.71 |
ENSDART00000171255
|
COMP
|
cartilage oligomeric matrix protein |
| chr20_+_27506658 | 2.70 |
|
|
|
| chr25_+_8279754 | 2.64 |
ENSDART00000073410
|
muc5.1
|
mucin 5.1, oligomeric mucus/gel-forming |
| chr12_-_22438379 | 2.63 |
ENSDART00000177715
|
CR925753.1
|
ENSDARG00000108493 |
| chr20_-_475417 | 2.59 |
ENSDART00000032212
|
fynrk
|
fyn-related Src family tyrosine kinase |
| chr24_-_33617592 | 2.56 |
|
|
|
| chr10_-_37131530 | 2.52 |
ENSDART00000132023
|
myo18aa
|
myosin XVIIIAa |
| chr21_-_39013447 | 2.51 |
ENSDART00000114885
|
aldh3a2b
|
aldehyde dehydrogenase 3 family, member A2b |
| chr22_+_520496 | 2.50 |
ENSDART00000067638
|
med20
|
mediator complex subunit 20 |
| chr1_+_9999665 | 2.50 |
ENSDART00000054879
|
ENSDARG00000037679
|
ENSDARG00000037679 |
| chr7_+_20272091 | 2.49 |
ENSDART00000052917
|
slc3a2a
|
solute carrier family 3 (amino acid transporter heavy chain), member 2a |
| chr2_-_30198789 | 2.40 |
ENSDART00000019149
|
rpl7
|
ribosomal protein L7 |
| chr22_+_5722410 | 2.39 |
ENSDART00000143052
|
cox14
|
COX14 cytochrome c oxidase assembly factor |
| chr20_-_4695108 | 2.35 |
ENSDART00000167224
ENSDART00000125620 ENSDART00000053858 |
papola
|
poly(A) polymerase alpha |
| chr3_-_55995924 | 2.34 |
ENSDART00000167356
|
tfap4
|
transcription factor AP-4 (activating enhancer binding protein 4) |
| chr12_-_36294319 | 2.33 |
ENSDART00000157746
ENSDART00000166925 |
pde6g
|
phosphodiesterase 6G, cGMP-specific, rod, gamma |
| chr16_-_2322075 | 2.32 |
|
|
|
| chr9_+_24089828 | 2.32 |
ENSDART00000127859
|
trim63b
|
tripartite motif containing 63b |
| chr10_-_24401876 | 2.26 |
ENSDART00000149362
|
pitpnab
|
phosphatidylinositol transfer protein, alpha b |
| chr7_-_31170266 | 2.23 |
ENSDART00000075398
|
cilp
|
cartilage intermediate layer protein, nucleotide pyrophosphohydrolase |
| chr9_-_32942783 | 2.23 |
ENSDART00000060006
|
olig2
|
oligodendrocyte lineage transcription factor 2 |
| chr14_-_27703812 | 2.22 |
ENSDART00000054115
|
tsc22d3
|
TSC22 domain family, member 3 |
| chr5_-_67233396 | 2.21 |
ENSDART00000051833
ENSDART00000124890 |
gsx1
|
GS homeobox 1 |
| chr19_-_5416317 | 2.19 |
ENSDART00000010373
|
krt1-19d
|
keratin, type 1, gene 19d |
| chr13_-_1177857 | 2.19 |
ENSDART00000111052
|
olig3
|
oligodendrocyte transcription factor 3 |
| chr2_+_1602501 | 2.17 |
ENSDART00000109668
|
bcl10
|
B-cell CLL/lymphoma 10 |
| chr25_+_30535271 | 2.16 |
|
|
|
| chr21_-_20305406 | 2.16 |
ENSDART00000065659
|
rbp4l
|
retinol binding protein 4, like |
| chr2_+_4233224 | 2.14 |
ENSDART00000163737
|
mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr5_-_63487161 | 2.07 |
ENSDART00000048395
|
cmlc1
|
cardiac myosin light chain-1 |
| chr14_-_45883839 | 2.06 |
|
|
|
| chr10_+_44364117 | 2.05 |
|
|
|
| chr19_+_42183946 | 2.04 |
ENSDART00000087096
|
tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr11_+_15478585 | 2.03 |
ENSDART00000163289
ENSDART00000066033 |
gdf11
|
growth differentiation factor 11 |
| chr15_+_895792 | 2.01 |
|
|
|
| chr11_+_33972923 | 2.00 |
ENSDART00000172594
|
fam43a
|
family with sequence similarity 43, member A |
| chr23_+_27052594 | 1.98 |
ENSDART00000109712
ENSDART00000018654 |
rnd1b
|
Rho family GTPase 1b |
| chr4_-_20764499 | 1.97 |
ENSDART00000023337
|
napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr22_-_11596329 | 1.94 |
ENSDART00000063133
|
gcga
|
glucagon a |
| chr5_-_40451061 | 1.94 |
ENSDART00000083515
|
pdzd2
|
PDZ domain containing 2 |
| chr7_+_73601671 | 1.94 |
ENSDART00000163075
|
zgc:173552
|
zgc:173552 |
| chr22_+_10372455 | 1.92 |
ENSDART00000122403
|
wu:fb55g09
|
wu:fb55g09 |
| chr14_-_32993686 | 1.92 |
ENSDART00000146999
|
rpl39
|
ribosomal protein L39 |
| chr24_-_25546068 | 1.91 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr3_+_5665300 | 1.91 |
ENSDART00000101807
|
si:ch211-106h11.3
|
si:ch211-106h11.3 |
| chr24_-_4117031 | 1.83 |
ENSDART00000147849
|
klf6a
|
Kruppel-like factor 6a |
| chr7_+_71658461 | 1.83 |
|
|
|
| chr4_-_71335661 | 1.81 |
|
|
|
| chr8_+_17042976 | 1.78 |
ENSDART00000023206
|
plk2b
|
polo-like kinase 2b (Drosophila) |
| chr25_+_34627031 | 1.76 |
ENSDART00000154377
|
HIST1H2BA (1 of many)
|
histone cluster 1 H2B family member a |
| chr19_-_44459390 | 1.76 |
ENSDART00000125168
|
uqcrb
|
ubiquinol-cytochrome c reductase binding protein |
| chr13_+_1398289 | 1.76 |
|
|
|
| chr12_-_19224698 | 1.74 |
ENSDART00000044860
|
maff
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr1_-_21987718 | 1.73 |
ENSDART00000128918
|
fgfbp1b
|
fibroblast growth factor binding protein 1b |
| chr3_-_36465046 | 1.71 |
ENSDART00000172003
|
si:dkeyp-72e1.6
|
si:dkeyp-72e1.6 |
| chr9_-_30453625 | 1.67 |
ENSDART00000060150
|
mid1ip1a
|
MID1 interacting protein 1a |
| chr23_-_10242235 | 1.67 |
ENSDART00000081223
ENSDART00000144280 |
krt5
|
keratin 5 |
| chr3_+_33209227 | 1.66 |
ENSDART00000128786
|
pyya
|
peptide YYa |
| chr5_+_8843643 | 1.62 |
ENSDART00000149417
ENSDART00000061616 |
tal2
|
T-cell acute lymphocytic leukemia 2 |
| chr23_+_18176720 | 1.59 |
ENSDART00000010270
|
mfsd4ab
|
major facilitator superfamily domain containing 4Ab |
| chr11_-_38940966 | 1.56 |
ENSDART00000105133
|
wnt4a
|
wingless-type MMTV integration site family, member 4a |
| chr11_-_42826276 | 1.55 |
|
|
|
| chr1_-_33981055 | 1.54 |
ENSDART00000102111
|
tpt1
|
tumor protein, translationally-controlled 1 |
| chr20_-_54192092 | 1.54 |
ENSDART00000023550
|
hsp90aa1.2
|
heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 2 |
| chr10_-_7827244 | 1.52 |
ENSDART00000111058
|
mpx
|
myeloid-specific peroxidase |
| chr13_+_30546398 | 1.51 |
|
|
|
| chr5_-_13184573 | 1.49 |
ENSDART00000018351
|
zgc:65851
|
zgc:65851 |
| chr2_-_59006559 | 1.48 |
|
|
|
| chr24_-_1308047 | 1.47 |
ENSDART00000164904
|
nrp1a
|
neuropilin 1a |
| chr21_+_10663517 | 1.46 |
ENSDART00000074833
|
rx3
|
retinal homeobox gene 3 |
| chr3_+_15355424 | 1.45 |
ENSDART00000141714
|
nupr1
|
nuclear protein 1 |
| chr7_-_35042805 | 1.43 |
ENSDART00000005053
|
slc12a4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr22_+_38028979 | 1.43 |
ENSDART00000165078
ENSDART00000159906 |
CU659306.1
|
ENSDARG00000100625 |
| chr18_-_16934961 | 1.41 |
ENSDART00000122102
|
wee1
|
WEE1 G2 checkpoint kinase |
| chr21_-_37924790 | 1.41 |
ENSDART00000131490
|
ripply1
|
ripply transcriptional repressor 1 |
| chr22_-_14247636 | 1.41 |
|
|
|
| chr24_+_37800773 | 1.40 |
ENSDART00000137017
|
h3f3d
|
H3 histone, family 3D |
| chr21_-_14276856 | 1.39 |
|
|
|
| chr3_+_19849761 | 1.39 |
|
|
|
| chr8_+_7237516 | 1.39 |
|
|
|
| chr7_-_13638486 | 1.35 |
ENSDART00000169828
|
rlbp1a
|
retinaldehyde binding protein 1a |
| chr10_-_22875997 | 1.35 |
ENSDART00000079454
|
vamp2
|
vesicle-associated membrane protein 2 |
| chr17_-_37266525 | 1.34 |
ENSDART00000128715
|
kif3cb
|
kinesin family member 3Cb |
| chr13_+_24417183 | 1.34 |
ENSDART00000177677
|
lgalslb
|
lectin, galactoside-binding-like b |
| chr21_+_32285006 | 1.33 |
|
|
|
| chr3_-_14348442 | 1.32 |
ENSDART00000172102
|
elof1
|
ELF1 homolog, elongation factor 1 |
| chr16_+_24045707 | 1.31 |
ENSDART00000058965
|
apoeb
|
apolipoprotein Eb |
| chr25_-_352985 | 1.31 |
ENSDART00000165705
|
szl
|
sizzled |
| chr8_-_6833841 | 1.31 |
ENSDART00000005321
|
neflb
|
neurofilament, light polypeptide b |
| chr19_+_20392669 | 1.30 |
ENSDART00000052627
|
npvf
|
neuropeptide VF precursor |
| chr21_+_43674754 | 1.29 |
ENSDART00000136025
|
tmlhe
|
trimethyllysine hydroxylase, epsilon |
| chr24_-_25545773 | 1.29 |
ENSDART00000015391
|
chrnd
|
cholinergic receptor, nicotinic, delta (muscle) |
| chr10_-_35015129 | 1.28 |
ENSDART00000156045
|
CT573139.2
|
ENSDARG00000098015 |
| chr13_-_2083873 | 1.28 |
ENSDART00000129773
|
mlip
|
muscular LMNA-interacting protein |
| chr1_+_28293053 | 1.24 |
ENSDART00000075539
|
cryaa
|
crystallin, alpha A |
| chr2_-_7560202 | 1.24 |
ENSDART00000164494
|
asip2b
|
agouti signaling protein, nonagouti homolog (mouse) 2b |
| chr7_+_73594790 | 1.23 |
ENSDART00000123081
|
zgc:173552
|
zgc:173552 |
| chr24_-_32281758 | 1.21 |
ENSDART00000151845
|
trdmt1
|
tRNA aspartic acid methyltransferase 1 |
| chr22_-_26307952 | 1.20 |
|
|
|
| chr22_-_10135735 | 1.20 |
ENSDART00000142012
|
rbck1
|
RanBP-type and C3HC4-type zinc finger containing 1 |
| chr2_+_24851910 | 1.19 |
ENSDART00000133109
|
rps28
|
ribosomal protein S28 |
| chr14_+_10350918 | 1.18 |
ENSDART00000125865
|
atrx
|
alpha thalassemia/mental retardation syndrome X-linked homolog (human) |
| chr11_-_5617162 | 1.18 |
ENSDART00000066168
|
pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr20_-_7141247 | 1.17 |
ENSDART00000160910
|
si:ch211-121a2.4
|
si:ch211-121a2.4 |
| chr4_+_73490746 | 1.17 |
ENSDART00000174082
|
nup50
|
nucleoporin 50 |
| chr2_+_43719715 | 1.17 |
ENSDART00000037808
|
itgb1b.2
|
integrin, beta 1b.2 |
| chr3_-_59179368 | 1.14 |
ENSDART00000100327
|
nptx1l
|
neuronal pentraxin 1 like |
| chr14_-_10311677 | 1.13 |
ENSDART00000133723
|
si:dkey-92i17.2
|
si:dkey-92i17.2 |
| chr6_-_34955205 | 1.12 |
ENSDART00000131610
|
serbp1a
|
SERPINE1 mRNA binding protein 1a |
| chr5_-_64396143 | 1.10 |
ENSDART00000168486
|
inpp5e
|
inositol polyphosphate-5-phosphatase E |
| chr13_-_40027211 | 1.07 |
|
|
|
| chr22_-_17570275 | 1.07 |
ENSDART00000099056
|
gpx4a
|
glutathione peroxidase 4a |
| chr24_-_24017962 | 1.05 |
ENSDART00000080602
|
map7d2b
|
MAP7 domain containing 2b |
| chr20_+_27121126 | 1.04 |
ENSDART00000126919
ENSDART00000016014 |
chga
|
chromogranin A |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.4 | 10.1 | GO:0019730 | antimicrobial humoral response(GO:0019730) antibacterial humoral response(GO:0019731) |
| 3.3 | 16.7 | GO:0065005 | plasma lipoprotein particle assembly(GO:0034377) high-density lipoprotein particle assembly(GO:0034380) protein-lipid complex assembly(GO:0065005) |
| 2.6 | 7.9 | GO:0006601 | creatine metabolic process(GO:0006600) creatine biosynthetic process(GO:0006601) |
| 2.0 | 5.9 | GO:0002824 | positive regulation of adaptive immune response(GO:0002821) positive regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains(GO:0002824) negative regulation of phospholipase activity(GO:0010519) positive regulation of vesicle fusion(GO:0031340) type 2 immune response(GO:0042092) regulation of fatty acid biosynthetic process(GO:0042304) T-helper 2 cell differentiation(GO:0045064) leukocyte apoptotic process(GO:0071887) regulation of leukocyte apoptotic process(GO:2000106) |
| 1.8 | 5.4 | GO:0060055 | larval development(GO:0002164) larval heart development(GO:0007508) angiogenesis involved in wound healing(GO:0060055) |
| 1.7 | 5.0 | GO:1904019 | epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) |
| 1.5 | 6.1 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 1.3 | 3.9 | GO:0003223 | ventricular compact myocardium morphogenesis(GO:0003223) |
| 1.0 | 5.1 | GO:0046677 | response to antibiotic(GO:0046677) |
| 1.0 | 4.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 1.0 | 5.9 | GO:0009136 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) |
| 0.9 | 2.8 | GO:0003085 | negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.9 | 3.6 | GO:0046888 | negative regulation of hormone secretion(GO:0046888) |
| 0.8 | 1.6 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) |
| 0.8 | 3.8 | GO:0070328 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.7 | 2.2 | GO:0021742 | abducens nucleus development(GO:0021742) |
| 0.7 | 48.5 | GO:0007601 | visual perception(GO:0007601) |
| 0.7 | 2.0 | GO:0070292 | N-acylphosphatidylethanolamine metabolic process(GO:0070292) |
| 0.6 | 5.2 | GO:0042396 | phosphagen metabolic process(GO:0006599) phosphocreatine metabolic process(GO:0006603) phosphagen biosynthetic process(GO:0042396) phosphocreatine biosynthetic process(GO:0046314) |
| 0.5 | 2.1 | GO:0061195 | dorsal spinal cord development(GO:0021516) tongue development(GO:0043586) tongue morphogenesis(GO:0043587) taste bud development(GO:0061193) taste bud morphogenesis(GO:0061194) taste bud formation(GO:0061195) cranial ganglion development(GO:0061550) |
| 0.5 | 4.6 | GO:0070309 | lens fiber cell development(GO:0070307) lens fiber cell morphogenesis(GO:0070309) |
| 0.5 | 10.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.5 | 1.5 | GO:0060898 | eye field cell fate commitment involved in camera-type eye formation(GO:0060898) |
| 0.5 | 3.2 | GO:0021627 | olfactory nerve development(GO:0021553) olfactory nerve morphogenesis(GO:0021627) olfactory nerve formation(GO:0021628) |
| 0.4 | 1.7 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.4 | 3.0 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.4 | 2.1 | GO:0055015 | ventricular cardiac muscle cell differentiation(GO:0055012) ventricular cardiac muscle cell development(GO:0055015) |
| 0.3 | 10.4 | GO:0060030 | dorsal convergence(GO:0060030) |
| 0.3 | 4.6 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.3 | 1.0 | GO:2000471 | regulation of hematopoietic stem cell migration(GO:2000471) |
| 0.3 | 2.5 | GO:0060142 | regulation of syncytium formation by plasma membrane fusion(GO:0060142) |
| 0.3 | 1.2 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.3 | 0.9 | GO:0003228 | atrial cardiac muscle tissue development(GO:0003228) |
| 0.3 | 2.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.3 | 1.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.2 | 2.4 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.2 | 4.5 | GO:0015807 | L-amino acid transport(GO:0015807) |
| 0.2 | 0.9 | GO:0045337 | farnesyl diphosphate biosynthetic process(GO:0045337) |
| 0.2 | 3.5 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.2 | 0.7 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.2 | 1.3 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.2 | 3.8 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.2 | 1.0 | GO:0031086 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.2 | 4.7 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 0.5 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.2 | 1.8 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.2 | 0.3 | GO:0048341 | paraxial mesoderm morphogenesis(GO:0048340) paraxial mesoderm formation(GO:0048341) |
| 0.2 | 3.7 | GO:0061035 | regulation of cartilage development(GO:0061035) |
| 0.1 | 1.7 | GO:0042743 | hydrogen peroxide metabolic process(GO:0042743) |
| 0.1 | 7.1 | GO:0007160 | cell-matrix adhesion(GO:0007160) |
| 0.1 | 12.4 | GO:0031101 | fin regeneration(GO:0031101) |
| 0.1 | 1.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 1.7 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.1 | 1.3 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.1 | 0.7 | GO:0030317 | sperm motility(GO:0030317) |
| 0.1 | 0.7 | GO:0006046 | N-acetylglucosamine catabolic process(GO:0006046) |
| 0.1 | 1.4 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 1.4 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 2.2 | GO:0051092 | positive regulation of NF-kappaB transcription factor activity(GO:0051092) |
| 0.1 | 0.4 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 2.4 | GO:0043631 | mRNA polyadenylation(GO:0006378) RNA polyadenylation(GO:0043631) |
| 0.1 | 1.9 | GO:0003323 | type B pancreatic cell development(GO:0003323) |
| 0.1 | 2.2 | GO:0030513 | positive regulation of BMP signaling pathway(GO:0030513) |
| 0.1 | 1.6 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) |
| 0.1 | 0.9 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.2 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.1 | 1.4 | GO:0007634 | optokinetic behavior(GO:0007634) |
| 0.1 | 2.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.1 | 0.8 | GO:0046579 | positive regulation of Ras protein signal transduction(GO:0046579) positive regulation of small GTPase mediated signal transduction(GO:0051057) |
| 0.1 | 2.4 | GO:0031018 | endocrine pancreas development(GO:0031018) |
| 0.1 | 1.1 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.1 | 0.3 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.1 | 5.7 | GO:0033993 | response to lipid(GO:0033993) |
| 0.1 | 2.5 | GO:0006081 | cellular aldehyde metabolic process(GO:0006081) |
| 0.1 | 1.3 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.8 | GO:0099515 | vesicle transport along actin filament(GO:0030050) actin filament-based transport(GO:0099515) |
| 0.0 | 0.9 | GO:0031124 | mRNA 3'-end processing(GO:0031124) |
| 0.0 | 3.2 | GO:0042391 | regulation of membrane potential(GO:0042391) |
| 0.0 | 0.4 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.9 | GO:0044773 | mitotic DNA damage checkpoint(GO:0044773) |
| 0.0 | 2.7 | GO:0045665 | negative regulation of neuron differentiation(GO:0045665) |
| 0.0 | 1.3 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 0.6 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.3 | GO:0042551 | neuron maturation(GO:0042551) |
| 0.0 | 1.8 | GO:0001756 | somitogenesis(GO:0001756) |
| 0.0 | 0.2 | GO:0048798 | swim bladder maturation(GO:0048796) swim bladder inflation(GO:0048798) |
| 0.0 | 2.0 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.0 | GO:0046619 | optic placode formation(GO:0001743) optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 1.2 | GO:0006606 | protein import into nucleus(GO:0006606) protein targeting to nucleus(GO:0044744) single-organism nuclear import(GO:1902593) |
| 0.0 | 2.0 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.6 | GO:0018108 | peptidyl-tyrosine phosphorylation(GO:0018108) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.7 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 6.8 | GO:0006412 | translation(GO:0006412) |
| 0.0 | 1.1 | GO:0006979 | response to oxidative stress(GO:0006979) |
| 0.0 | 2.0 | GO:0009952 | anterior/posterior pattern specification(GO:0009952) |
| 0.0 | 0.0 | GO:0065001 | specification of organ axis polarity(GO:0010084) specification of axis polarity(GO:0065001) |
| 0.0 | 0.3 | GO:0007007 | inner mitochondrial membrane organization(GO:0007007) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.8 | GO:0098556 | cytoplasmic side of rough endoplasmic reticulum membrane(GO:0098556) |
| 1.9 | 7.5 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 1.8 | 9.2 | GO:0042627 | chylomicron(GO:0042627) |
| 1.5 | 4.6 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 1.1 | 5.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.7 | 5.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.4 | 5.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.4 | 3.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.3 | 7.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.3 | 15.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.2 | 11.7 | GO:0016459 | myosin complex(GO:0016459) |
| 0.2 | 0.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.2 | 0.9 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.2 | 1.3 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.2 | 8.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.2 | 1.8 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.2 | 7.1 | GO:0022626 | cytosolic large ribosomal subunit(GO:0022625) cytosolic ribosome(GO:0022626) |
| 0.1 | 1.2 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 0.9 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 3.4 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 8.9 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.1 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 2.1 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 2.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 7.7 | GO:0031012 | extracellular matrix(GO:0031012) |
| 0.0 | 1.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.3 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 3.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.0 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.6 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 6.8 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.4 | GO:0098798 | mitochondrial protein complex(GO:0098798) |
| 0.0 | 1.2 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 4.7 | GO:0005764 | lysosome(GO:0005764) |
| 0.0 | 2.4 | GO:0031966 | mitochondrial membrane(GO:0031966) |
| 0.0 | 10.8 | GO:0005576 | extracellular region(GO:0005576) |
| 0.0 | 1.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 2.0 | GO:0005938 | cell cortex(GO:0005938) |
| 0.0 | 0.9 | GO:0016591 | DNA-directed RNA polymerase II, holoenzyme(GO:0016591) |
| 0.0 | 0.2 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 7.5 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 1.7 | 48.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.5 | 9.2 | GO:0071814 | lipoprotein particle binding(GO:0071813) protein-lipid complex binding(GO:0071814) |
| 1.5 | 6.1 | GO:0019809 | polyamine binding(GO:0019808) spermidine binding(GO:0019809) |
| 1.3 | 3.8 | GO:0070513 | death domain binding(GO:0070513) |
| 0.8 | 7.0 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.7 | 5.9 | GO:0055102 | phospholipase inhibitor activity(GO:0004859) lipase inhibitor activity(GO:0055102) |
| 0.7 | 4.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.6 | 5.2 | GO:0016775 | creatine kinase activity(GO:0004111) phosphotransferase activity, nitrogenous group as acceptor(GO:0016775) |
| 0.6 | 5.4 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.6 | 5.9 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.6 | 2.4 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.5 | 1.6 | GO:0016743 | carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.5 | 3.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.5 | 2.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.4 | 2.2 | GO:0034632 | retinol transporter activity(GO:0034632) |
| 0.4 | 4.7 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.4 | 1.3 | GO:0005521 | lamin binding(GO:0005521) |
| 0.4 | 1.2 | GO:0098634 | C-X3-C chemokine binding(GO:0019960) protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.4 | 7.9 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.4 | 1.1 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 0.3 | 2.0 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.3 | 3.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.3 | 2.5 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.3 | 2.9 | GO:0005344 | oxygen transporter activity(GO:0005344) oxygen binding(GO:0019825) |
| 0.2 | 1.6 | GO:0015145 | glucose transmembrane transporter activity(GO:0005355) monosaccharide transmembrane transporter activity(GO:0015145) hexose transmembrane transporter activity(GO:0015149) |
| 0.2 | 2.9 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.2 | 4.5 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.1 | 1.4 | GO:0015377 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 0.9 | GO:0016493 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 1.0 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.1 | 0.9 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 0.4 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.1 | 21.1 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.1 | 13.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.1 | 2.2 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.1 | 0.7 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.1 | 1.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.4 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 2.8 | GO:0051427 | hormone receptor binding(GO:0051427) |
| 0.1 | 1.7 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.1 | 0.5 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 1.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.1 | 1.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.5 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 4.7 | GO:0030414 | peptidase inhibitor activity(GO:0030414) |
| 0.0 | 1.0 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 25.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 1.7 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 3.8 | GO:0004857 | enzyme inhibitor activity(GO:0004857) |
| 0.0 | 0.7 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 9.5 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 15.6 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.0 | 1.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 2.1 | GO:0042802 | identical protein binding(GO:0042802) |
| 0.0 | 1.3 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 1.3 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.0 | 0.4 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.0 | 1.4 | GO:1902936 | phosphatidylinositol bisphosphate binding(GO:1902936) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 2.6 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 1.0 | GO:0018024 | histone-lysine N-methyltransferase activity(GO:0018024) |
| 0.0 | 0.7 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 1.9 | GO:0001664 | G-protein coupled receptor binding(GO:0001664) |
| 0.0 | 1.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 1.1 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 2.0 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 4.6 | GO:0003779 | actin binding(GO:0003779) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.2 | 3.7 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.2 | 2.3 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.2 | 7.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 3.9 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 7.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 1.0 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.1 | 1.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 2.1 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 1.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.9 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.0 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.5 | 3.2 | REACTOME ACETYLCHOLINE BINDING AND DOWNSTREAM EVENTS | Genes involved in Acetylcholine Binding And Downstream Events |
| 0.4 | 1.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.4 | 5.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.3 | 14.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.3 | 2.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.2 | 2.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.2 | 10.6 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.2 | 2.1 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.2 | 2.9 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 1.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 3.4 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 2.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.1 | 2.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 6.3 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.1 | 4.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.1 | 0.6 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 0.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 1.0 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 1.2 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 3.4 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 4.8 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 1.8 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.0 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 0.7 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.2 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |