Project
DANIO-CODE
Navigation
Downloads
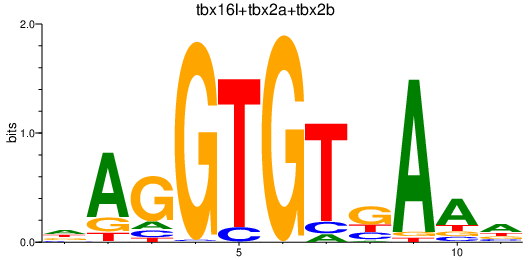
Results for tbx16l+tbx2a+tbx2b
Z-value: 1.33
Transcription factors associated with tbx16l+tbx2a+tbx2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbx2b
|
ENSDARG00000006120 | T-box transcription factor 2b |
|
tbx16l
|
ENSDARG00000006939 | T-box transcription factor 16, like |
|
tbx2a
|
ENSDARG00000018025 | T-box transcription factor 2a |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbx2a | dr10_dc_chr5_-_55843553_55843669 | -0.68 | 3.7e-03 | Click! |
| tbx2b | dr10_dc_chr15_+_27431469_27431569 | -0.55 | 2.7e-02 | Click! |
| tbx6l | dr10_dc_chr5_+_41680352_41680459 | 0.04 | 8.8e-01 | Click! |
Activity profile of tbx16l+tbx2a+tbx2b motif
Sorted Z-values of tbx16l+tbx2a+tbx2b motif
Network of associatons between targets according to the STRING database.
First level regulatory network of tbx16l+tbx2a+tbx2b
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_+_1283662 | 3.84 |
ENSDART00000124161
|
si:ch73-138e16.5
|
si:ch73-138e16.5 |
| chr2_-_53772219 | 3.80 |
ENSDART00000037557
|
admp
|
anti-dorsalizing morphogenic protein |
| chr2_+_34984631 | 3.22 |
ENSDART00000141796
|
astn1
|
astrotactin 1 |
| chr1_-_48876924 | 2.96 |
ENSDART00000143474
|
zp3c
|
zona pellucida glycoprotein 3c |
| chr15_-_1625846 | 2.92 |
ENSDART00000081875
|
nnr
|
nanor |
| chr17_+_26946957 | 2.86 |
ENSDART00000114215
|
grhl3
|
grainyhead-like transcription factor 3 |
| chr23_+_23305376 | 2.55 |
ENSDART00000126479
|
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr10_-_1933874 | 2.52 |
ENSDART00000101023
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr17_-_20264762 | 2.49 |
|
|
|
| chr14_-_33641145 | 2.43 |
ENSDART00000167774
|
foxa
|
forkhead box A sequence |
| chr19_-_35411993 | 2.34 |
ENSDART00000051751
|
zgc:113424
|
zgc:113424 |
| chr23_+_23305483 | 2.27 |
ENSDART00000126479
|
plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr7_-_58523645 | 2.17 |
|
|
|
| chr17_-_20264870 | 2.17 |
|
|
|
| chr7_+_23799153 | 2.14 |
|
|
|
| chr11_-_25019899 | 2.13 |
ENSDART00000123567
|
snai1a
|
snail family zinc finger 1a |
| chr11_+_29299382 | 2.11 |
|
|
|
| chr12_+_29121368 | 2.08 |
ENSDART00000006505
|
mxtx2
|
mix-type homeobox gene 2 |
| chr17_-_8155740 | 2.05 |
ENSDART00000064678
|
lft2
|
lefty2 |
| chr6_-_2000017 | 2.05 |
ENSDART00000158535
|
vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr5_+_60791165 | 1.91 |
ENSDART00000050906
|
lrwd1
|
leucine-rich repeats and WD repeat domain containing 1 |
| chr12_-_14104939 | 1.91 |
ENSDART00000152742
|
buc2l
|
bucky ball 2-like |
| chr22_-_8767667 | 1.77 |
ENSDART00000148005
|
CR450686.2
|
ENSDARG00000093787 |
| chr2_+_30199024 | 1.76 |
ENSDART00000004903
|
rdh10b
|
retinol dehydrogenase 10b |
| chr20_-_43826667 | 1.76 |
ENSDART00000100637
|
mixl1
|
Mix paired-like homeobox |
| chr11_-_25019298 | 1.73 |
ENSDART00000130477
ENSDART00000079578 |
snai1a
|
snail family zinc finger 1a |
| chr20_-_14218080 | 1.72 |
ENSDART00000104032
|
si:ch211-223m11.2
|
si:ch211-223m11.2 |
| chr20_-_34894930 | 1.71 |
|
|
|
| chr24_+_9887575 | 1.69 |
ENSDART00000172773
|
si:ch211-146l10.8
|
si:ch211-146l10.8 |
| chr22_-_36721626 | 1.67 |
ENSDART00000017188
ENSDART00000124698 |
ncl
|
nucleolin |
| chr7_+_13742576 | 1.62 |
|
|
|
| chr3_-_23513177 | 1.59 |
ENSDART00000078425
|
eve1
|
even-skipped-like1 |
| chr3_-_22240424 | 1.59 |
|
|
|
| chr21_-_9291200 | 1.58 |
ENSDART00000160932
|
sdad1
|
SDA1 domain containing 1 |
| chr25_+_29219700 | 1.53 |
ENSDART00000108692
ENSDART00000077445 |
pim3
|
Pim-3 proto-oncogene, serine/threonine kinase |
| KN150589v1_-_6303 | 1.53 |
|
|
|
| chr1_-_51870424 | 1.51 |
ENSDART00000135636
|
acy3.1
|
aspartoacylase (aminocyclase) 3, tandem duplicate 1 |
| chr24_+_32295105 | 1.48 |
ENSDART00000143570
|
CU207275.1
|
ENSDARG00000092965 |
| chr2_+_30198819 | 1.48 |
ENSDART00000004903
|
rdh10b
|
retinol dehydrogenase 10b |
| chr15_-_17163526 | 1.48 |
ENSDART00000101724
|
mos
|
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr4_+_17653548 | 1.47 |
ENSDART00000026509
|
cwf19l1
|
CWF19-like 1, cell cycle control |
| chr7_+_51520610 | 1.47 |
ENSDART00000174201
|
slc38a7
|
solute carrier family 38, member 7 |
| chr14_+_50263527 | 1.46 |
ENSDART00000126260
|
anxa6
|
annexin A6 |
| chr22_+_25714282 | 1.46 |
ENSDART00000143367
|
si:dkeyp-98a7.9
|
si:dkeyp-98a7.9 |
| chr5_+_29052293 | 1.45 |
ENSDART00000035400
|
tsc1a
|
tuberous sclerosis 1a |
| chr10_+_15644868 | 1.44 |
ENSDART00000139259
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr21_-_3548863 | 1.44 |
ENSDART00000086492
|
atp8b1
|
ATPase, aminophospholipid transporter, class I, type 8B, member 1 |
| chr13_-_50299643 | 1.44 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr7_+_36196295 | 1.43 |
ENSDART00000146202
|
aktip
|
akt interacting protein |
| chr5_+_61711614 | 1.42 |
ENSDART00000082965
|
ABR
|
active BCR-related |
| chr11_-_40383013 | 1.42 |
ENSDART00000112140
|
fam213b
|
family with sequence similarity 213, member B |
| chr12_-_8920454 | 1.39 |
ENSDART00000041728
|
cyp26a1
|
cytochrome P450, family 26, subfamily A, polypeptide 1 |
| chr14_-_30200514 | 1.38 |
ENSDART00000026907
ENSDART00000126216 |
her11
|
hairy-related 11 |
| chr18_-_11626694 | 1.37 |
ENSDART00000098565
|
CRACR2A
|
calcium release activated channel regulator 2A |
| chr9_+_8402397 | 1.36 |
ENSDART00000133501
|
si:ch1073-75o15.4
|
si:ch1073-75o15.4 |
| chr11_-_3516034 | 1.34 |
ENSDART00000009788
|
ddx19
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 19 (DBP5 homolog, yeast) |
| chr17_+_34238753 | 1.33 |
ENSDART00000014306
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr11_-_11591351 | 1.32 |
ENSDART00000142208
|
zgc:110712
|
zgc:110712 |
| chr7_+_15081529 | 1.30 |
ENSDART00000006018
|
mespaa
|
mesoderm posterior aa |
| chr8_+_3372903 | 1.30 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr12_+_6197452 | 1.29 |
|
|
|
| chr1_+_33962885 | 1.29 |
ENSDART00000145535
|
gtf2f2a
|
general transcription factor IIF, polypeptide 2a |
| chr2_-_53771952 | 1.28 |
ENSDART00000037557
|
admp
|
anti-dorsalizing morphogenic protein |
| chr21_+_20974216 | 1.27 |
ENSDART00000079692
|
ndr1
|
nodal-related 1 |
| KN150339v1_-_39357 | 1.26 |
ENSDART00000169638
|
wu:fa19b12
|
wu:fa19b12 |
| chr21_-_9633519 | 1.24 |
|
|
|
| chr20_-_34894607 | 1.23 |
|
|
|
| chr3_-_25142301 | 1.23 |
ENSDART00000145315
|
ddx5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr25_-_17271237 | 1.22 |
ENSDART00000110692
|
cyp2x6
|
cytochrome P450, family 2, subfamily X, polypeptide 6 |
| chr11_-_25019165 | 1.22 |
ENSDART00000130477
ENSDART00000079578 |
snai1a
|
snail family zinc finger 1a |
| chr5_-_15971338 | 1.22 |
ENSDART00000110437
|
piwil2
|
piwi-like RNA-mediated gene silencing 2 |
| chr13_-_9110887 | 1.21 |
ENSDART00000138833
|
grxcr1
|
glutaredoxin, cysteine rich 1 |
| chr10_-_1933761 | 1.21 |
ENSDART00000101023
|
tdgf1
|
teratocarcinoma-derived growth factor 1 |
| chr5_+_29052517 | 1.19 |
ENSDART00000035400
|
tsc1a
|
tuberous sclerosis 1a |
| chr25_-_24440001 | 1.19 |
ENSDART00000156805
|
CR854832.1
|
ENSDARG00000096817 |
| chr12_-_8920541 | 1.18 |
ENSDART00000041728
|
cyp26a1
|
cytochrome P450, family 26, subfamily A, polypeptide 1 |
| chr13_-_36409205 | 1.18 |
ENSDART00000043312
|
srsf5a
|
serine/arginine-rich splicing factor 5a |
| chr17_+_26946875 | 1.18 |
ENSDART00000114215
|
grhl3
|
grainyhead-like transcription factor 3 |
| chr5_-_54097015 | 1.17 |
|
|
|
| chr13_-_50299821 | 1.17 |
ENSDART00000170527
|
vent
|
ventral expressed homeobox |
| chr22_-_9854217 | 1.15 |
|
|
|
| chr20_+_35305119 | 1.14 |
ENSDART00000045135
|
fbxo16
|
F-box protein 16 |
| chr1_-_21021029 | 1.13 |
ENSDART00000129066
|
zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr6_+_8417443 | 1.13 |
|
|
|
| chr10_+_11014479 | 1.13 |
ENSDART00000064992
|
cdc37l1
|
cell division cycle 37-like 1 |
| chr7_+_56076854 | 1.12 |
|
|
|
| chr25_+_7261050 | 1.12 |
ENSDART00000163017
|
prc1a
|
protein regulator of cytokinesis 1a |
| chr20_+_26495546 | 1.11 |
ENSDART00000036942
|
zbtb2b
|
zinc finger and BTB domain containing 2b |
| chr19_-_3947422 | 1.11 |
ENSDART00000172271
|
thrap3b
|
thyroid hormone receptor associated protein 3b |
| chr9_+_55395128 | 1.10 |
ENSDART00000029528
|
mospd2
|
motile sperm domain containing 2 |
| chr7_-_52847458 | 1.10 |
ENSDART00000172179
ENSDART00000167882 |
cdh1
|
cadherin 1, type 1, E-cadherin (epithelial) |
| chr15_+_42440802 | 1.10 |
ENSDART00000089694
|
tiam1b
|
T-cell lymphoma invasion and metastasis 1b |
| chr15_-_25333773 | 1.08 |
ENSDART00000078230
|
mettl16
|
methyltransferase like 16 |
| chr15_-_29079613 | 1.07 |
ENSDART00000060018
|
dharma
|
dharma |
| chr8_+_3373066 | 1.07 |
ENSDART00000017850
|
ctu1
|
cytosolic thiouridylase subunit 1 homolog (S. pombe) |
| chr24_-_34794538 | 1.06 |
ENSDART00000171009
ENSDART00000170046 |
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr10_-_31129217 | 1.06 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr6_-_50588444 | 1.06 |
ENSDART00000151500
|
mtss1
|
metastasis suppressor 1 |
| chr12_-_46943717 | 1.06 |
ENSDART00000084557
|
lhpp
|
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
| chr7_-_18256512 | 1.05 |
ENSDART00000173539
|
rgs12a
|
regulator of G protein signaling 12a |
| chr25_-_31547164 | 1.04 |
ENSDART00000110180
|
duox2
|
dual oxidase 2 |
| chr24_+_40321763 | 1.03 |
|
|
|
| chr13_-_27223507 | 1.02 |
ENSDART00000101479
|
ddx43
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 43 |
| chr13_-_9110703 | 1.02 |
ENSDART00000138833
|
grxcr1
|
glutaredoxin, cysteine rich 1 |
| chr3_-_25143713 | 1.02 |
|
|
|
| chr21_+_26954898 | 1.01 |
ENSDART00000114469
|
fkbp2
|
FK506 binding protein 2 |
| chr12_+_18784456 | 1.01 |
ENSDART00000105854
|
josd1
|
Josephin domain containing 1 |
| chr5_+_50464723 | 1.01 |
ENSDART00000092002
|
pomt1
|
protein-O-mannosyltransferase 1 |
| chr13_-_7972442 | 1.00 |
ENSDART00000128005
|
atl2
|
atlastin GTPase 2 |
| chr5_+_58936399 | 1.00 |
|
|
|
| chr10_-_44619365 | 0.99 |
ENSDART00000085556
|
hip1ra
|
huntingtin interacting protein 1 related a |
| chr4_+_17653739 | 0.99 |
ENSDART00000026509
|
cwf19l1
|
CWF19-like 1, cell cycle control |
| chr20_-_23949621 | 0.99 |
|
|
|
| chr14_-_5510157 | 0.99 |
ENSDART00000131820
|
kazald2
|
Kazal-type serine peptidase inhibitor domain 2 |
| chr8_+_14949229 | 0.98 |
ENSDART00000045038
|
fnbp1l
|
formin binding protein 1-like |
| chr22_-_36721592 | 0.98 |
ENSDART00000130171
|
ncl
|
nucleolin |
| chr13_-_21541531 | 0.98 |
ENSDART00000067537
|
elovl6l
|
ELOVL family member 6, elongation of long chain fatty acids like |
| chr22_-_26537845 | 0.98 |
|
|
|
| chr3_-_8006 | 0.98 |
|
|
|
| chr23_-_18105560 | 0.96 |
|
|
|
| chr11_-_11925801 | 0.96 |
|
|
|
| chr5_+_26925238 | 0.96 |
ENSDART00000051491
|
sfrp1a
|
secreted frizzled-related protein 1a |
| chr8_-_19215929 | 0.95 |
ENSDART00000147172
|
abhd17ab
|
abhydrolase domain containing 17Ab |
| chr18_+_47305642 | 0.95 |
ENSDART00000009775
|
rbm7
|
RNA binding motif protein 7 |
| chr3_+_4123775 | 0.95 |
ENSDART00000056035
|
ENSDARG00000038398
|
ENSDARG00000038398 |
| chr7_+_38044801 | 0.94 |
ENSDART00000160197
|
BX323855.1
|
ENSDARG00000098940 |
| chr13_-_15662599 | 0.94 |
ENSDART00000145914
ENSDART00000010286 |
bag5
|
BCL2-associated athanogene 5 |
| chr10_-_210594 | 0.94 |
ENSDART00000059476
|
psmg1
|
proteasome (prosome, macropain) assembly chaperone 1 |
| chr10_-_31129270 | 0.93 |
ENSDART00000146116
|
panx3
|
pannexin 3 |
| chr8_-_38284748 | 0.93 |
ENSDART00000102233
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr1_+_29919378 | 0.93 |
ENSDART00000127670
|
bora
|
bora, aurora kinase A activator |
| chr17_+_31722625 | 0.93 |
ENSDART00000156180
|
arhgap5
|
Rho GTPase activating protein 5 |
| chr7_+_13742622 | 0.92 |
|
|
|
| chr15_+_42440952 | 0.92 |
ENSDART00000089694
|
tiam1b
|
T-cell lymphoma invasion and metastasis 1b |
| chr21_-_9291452 | 0.92 |
ENSDART00000169275
|
sdad1
|
SDA1 domain containing 1 |
| chr13_-_7972530 | 0.92 |
ENSDART00000080460
|
atl2
|
atlastin GTPase 2 |
| chr7_+_38408691 | 0.91 |
ENSDART00000171691
|
psmc3
|
proteasome 26S subunit, ATPase 3 |
| chr6_-_19812488 | 0.91 |
ENSDART00000151179
|
si:dkey-19e4.5
|
si:dkey-19e4.5 |
| chr21_-_9291682 | 0.91 |
ENSDART00000169275
|
sdad1
|
SDA1 domain containing 1 |
| chr6_+_20595096 | 0.90 |
|
|
|
| chr5_+_66693263 | 0.90 |
ENSDART00000142156
|
sebox
|
SEBOX homeobox |
| chr25_+_29219342 | 0.90 |
ENSDART00000108692
ENSDART00000077445 |
pim3
|
Pim-3 proto-oncogene, serine/threonine kinase |
| chr4_+_15007453 | 0.89 |
ENSDART00000146156
|
zc3hc1
|
zinc finger, C3HC-type containing 1 |
| chr1_-_52642562 | 0.89 |
ENSDART00000150579
|
znf330
|
zinc finger protein 330 |
| chr15_-_21733882 | 0.89 |
ENSDART00000156995
|
sorl1
|
sortilin-related receptor, L(DLR class) A repeats containing |
| chr7_+_15065241 | 0.89 |
ENSDART00000045385
|
mespba
|
mesoderm posterior ba |
| chr2_+_56534374 | 0.89 |
ENSDART00000113964
|
POLR2E (1 of many)
|
RNA polymerase II subunit E |
| chr7_-_58524285 | 0.88 |
ENSDART00000171095
|
sox32
|
SRY (sex determining region Y)-box 32 |
| chr13_-_28286577 | 0.88 |
ENSDART00000140989
|
BX950184.1
|
ENSDARG00000093030 |
| chr22_-_4414254 | 0.87 |
ENSDART00000114465
|
zgc:195170
|
zgc:195170 |
| chr25_+_32770756 | 0.86 |
ENSDART00000059800
|
lactb
|
lactamase, beta |
| chr17_+_15666078 | 0.86 |
ENSDART00000156726
|
bach2a
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2a |
| chr2_-_31703111 | 0.86 |
ENSDART00000126177
|
e2f5
|
E2F transcription factor 5 |
| chr9_+_30822402 | 0.86 |
|
|
|
| chr6_-_8225364 | 0.85 |
|
|
|
| chr15_+_44160948 | 0.85 |
ENSDART00000110060
|
zgc:165514
|
zgc:165514 |
| chr11_+_37345493 | 0.84 |
ENSDART00000157993
|
hp1bp3
|
heterochromatin protein 1, binding protein 3 |
| chr4_-_13974345 | 0.84 |
ENSDART00000067177
ENSDART00000144127 |
prickle1b
|
prickle homolog 1b |
| chr1_+_157840 | 0.84 |
ENSDART00000152205
ENSDART00000160843 |
cul4a
|
cullin 4A |
| chr5_+_25408083 | 0.83 |
|
|
|
| chr3_-_40090673 | 0.82 |
ENSDART00000155969
|
flii
|
flightless I actin binding protein |
| chr22_-_20378023 | 0.82 |
ENSDART00000010048
|
map2k2a
|
mitogen-activated protein kinase kinase 2a |
| chr21_+_39417430 | 0.81 |
ENSDART00000123970
|
mntb
|
MAX network transcriptional repressor b |
| chr20_+_5022501 | 0.81 |
ENSDART00000162678
|
si:dkey-60a16.1
|
si:dkey-60a16.1 |
| chr11_-_11591394 | 0.81 |
ENSDART00000142208
|
zgc:110712
|
zgc:110712 |
| chr20_+_43794079 | 0.80 |
ENSDART00000045185
|
lin9
|
lin-9 DREAM MuvB core complex component |
| chr24_-_34794856 | 0.80 |
ENSDART00000171009
ENSDART00000170046 |
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr20_-_38030337 | 0.80 |
ENSDART00000153451
|
angel2
|
angel homolog 2 (Drosophila) |
| chr1_-_48983245 | 0.80 |
|
|
|
| chr8_+_23083842 | 0.79 |
ENSDART00000025171
|
ythdf1
|
YTH N(6)-methyladenosine RNA binding protein 1 |
| chr7_+_16771283 | 0.79 |
ENSDART00000171275
|
CU929054.1
|
ENSDARG00000102605 |
| chr18_+_18077901 | 0.79 |
|
|
|
| chr1_+_49171052 | 0.78 |
ENSDART00000164936
|
lef1
|
lymphoid enhancer-binding factor 1 |
| chr7_+_20651600 | 0.78 |
ENSDART00000129161
|
wrap53
|
WD repeat containing, antisense to TP53 |
| chr25_+_32770866 | 0.78 |
ENSDART00000059800
|
lactb
|
lactamase, beta |
| chr24_-_34794463 | 0.78 |
ENSDART00000171009
ENSDART00000170046 |
ctnna1
|
catenin (cadherin-associated protein), alpha 1 |
| chr7_-_19989384 | 0.78 |
ENSDART00000173619
|
si:ch73-335l21.4
|
si:ch73-335l21.4 |
| chr7_+_41033439 | 0.78 |
ENSDART00000016660
|
ENSDARG00000016964
|
ENSDARG00000016964 |
| chr5_+_36168475 | 0.78 |
ENSDART00000146854
|
mark4a
|
MAP/microtubule affinity-regulating kinase 4a |
| chr4_-_12792010 | 0.77 |
ENSDART00000067131
|
irak3
|
interleukin-1 receptor-associated kinase 3 |
| chr15_-_104637 | 0.77 |
ENSDART00000164323
ENSDART00000161218 |
cyp2y3
|
cytochrome P450, family 2, subfamily Y, polypeptide 3 |
| chr6_+_37323636 | 0.77 |
ENSDART00000178969
ENSDART00000104180 |
zranb2
|
zinc finger, RAN-binding domain containing 2 |
| chr4_+_9279256 | 0.76 |
ENSDART00000014897
|
srgap1b
|
SLIT-ROBO Rho GTPase activating protein 1b |
| chr25_-_22791921 | 0.76 |
ENSDART00000128250
|
mob2a
|
MOB kinase activator 2a |
| chr15_+_42441012 | 0.76 |
ENSDART00000089694
|
tiam1b
|
T-cell lymphoma invasion and metastasis 1b |
| chr6_-_12787256 | 0.75 |
ENSDART00000157139
|
tmbim1a
|
transmembrane BAX inhibitor motif containing 1a |
| chr6_-_19473903 | 0.75 |
ENSDART00000126186
|
psmd12
|
proteasome 26S subunit, non-ATPase 12 |
| chr19_+_13457355 | 0.75 |
ENSDART00000172559
ENSDART00000163093 |
lrp12
|
low density lipoprotein receptor-related protein 12 |
| chr12_+_35855929 | 0.75 |
|
|
|
| chr6_-_7094468 | 0.75 |
|
|
|
| chr10_-_42931440 | 0.75 |
ENSDART00000123608
|
zgc:100918
|
zgc:100918 |
| chr22_-_37861903 | 0.75 |
ENSDART00000085931
|
acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr14_-_5510256 | 0.74 |
ENSDART00000131820
|
kazald2
|
Kazal-type serine peptidase inhibitor domain 2 |
| chr15_-_41849769 | 0.74 |
ENSDART00000156819
|
slc25a36b
|
solute carrier family 25 (pyrimidine nucleotide carrier ), member 36b |
| chr5_-_39136238 | 0.74 |
ENSDART00000127123
|
rasgef1ba
|
RasGEF domain family, member 1Ba |
| chr13_-_24778352 | 0.74 |
|
|
|
| chr15_-_43283562 | 0.74 |
ENSDART00000110352
|
ap1s3a
|
adaptor-related protein complex 1, sigma 3 subunit, a |
| chr5_+_37144525 | 0.73 |
ENSDART00000148766
|
dpf2
|
D4, zinc and double PHD fingers family 2 |
| chr3_-_7926 | 0.73 |
|
|
|
| chr5_-_39136156 | 0.73 |
ENSDART00000127123
|
rasgef1ba
|
RasGEF domain family, member 1Ba |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0072045 | convergent extension involved in nephron morphogenesis(GO:0072045) |
| 1.2 | 4.8 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.9 | 2.6 | GO:0071299 | mesodermal-endodermal cell signaling(GO:0003131) response to vitamin A(GO:0033189) cellular response to vitamin A(GO:0071299) retinoic acid receptor signaling pathway involved in somitogenesis(GO:0090242) |
| 0.7 | 5.1 | GO:0060063 | Spemann organizer formation at the embryonic shield(GO:0060063) |
| 0.5 | 2.2 | GO:0097101 | blood vessel endothelial cell fate specification(GO:0097101) |
| 0.5 | 1.1 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.5 | 3.2 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.5 | 1.5 | GO:0090083 | inclusion body assembly(GO:0070841) regulation of inclusion body assembly(GO:0090083) |
| 0.5 | 2.0 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.4 | 4.0 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.4 | 3.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.4 | 4.6 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) |
| 0.4 | 1.2 | GO:1990511 | piRNA biosynthetic process(GO:1990511) |
| 0.4 | 1.5 | GO:0001778 | plasma membrane repair(GO:0001778) chondrocyte differentiation involved in endochondral bone morphogenesis(GO:0003413) growth plate cartilage chondrocyte differentiation(GO:0003418) |
| 0.4 | 3.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.4 | 1.4 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.4 | 2.1 | GO:0060784 | regulation of cell proliferation involved in tissue homeostasis(GO:0060784) |
| 0.3 | 1.2 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.3 | 3.4 | GO:1900077 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.3 | 1.9 | GO:0071169 | establishment of protein localization to chromosome(GO:0070199) establishment of protein localization to chromatin(GO:0071169) |
| 0.3 | 0.5 | GO:0034087 | establishment of mitotic sister chromatid cohesion(GO:0034087) |
| 0.3 | 3.4 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.3 | 1.0 | GO:0021547 | midbrain-hindbrain boundary initiation(GO:0021547) |
| 0.3 | 0.8 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.3 | 4.4 | GO:2000344 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) egg coat formation(GO:0035803) regulation of acrosome reaction(GO:0060046) positive regulation of acrosome reaction(GO:2000344) |
| 0.3 | 1.3 | GO:1905132 | meiotic cell cycle phase transition(GO:0044771) metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.2 | 2.6 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.2 | 2.6 | GO:0061157 | RNA destabilization(GO:0050779) mRNA destabilization(GO:0061157) |
| 0.2 | 1.4 | GO:0030576 | nuclear body organization(GO:0030575) Cajal body organization(GO:0030576) |
| 0.2 | 0.9 | GO:0003262 | endocardial progenitor cell migration to the midline involved in heart field formation(GO:0003262) |
| 0.2 | 1.1 | GO:0003379 | establishment of cell polarity involved in gastrulation cell migration(GO:0003379) |
| 0.2 | 0.6 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.2 | 5.1 | GO:0008078 | mesodermal cell migration(GO:0008078) |
| 0.2 | 0.8 | GO:0015878 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) coenzyme transport(GO:0051182) |
| 0.2 | 0.6 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.2 | 0.6 | GO:0071047 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) nuclear polyadenylation-dependent mRNA catabolic process(GO:0071042) polyadenylation-dependent mRNA catabolic process(GO:0071047) |
| 0.2 | 0.9 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.2 | 0.7 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.2 | 0.7 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.2 | 0.6 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.2 | 0.7 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.2 | 3.0 | GO:0070593 | dendrite self-avoidance(GO:0070593) |
| 0.2 | 1.4 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.2 | 0.9 | GO:0021698 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) |
| 0.2 | 2.1 | GO:0003128 | heart field specification(GO:0003128) |
| 0.2 | 0.5 | GO:0071908 | determination of intestine left/right asymmetry(GO:0071908) |
| 0.1 | 0.6 | GO:0031338 | regulation of vesicle fusion(GO:0031338) maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.1 | 1.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.1 | 1.7 | GO:0098734 | protein depalmitoylation(GO:0002084) macromolecule depalmitoylation(GO:0098734) |
| 0.1 | 0.5 | GO:0042373 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) vitamin K metabolic process(GO:0042373) |
| 0.1 | 1.3 | GO:0031128 | developmental induction(GO:0031128) Spemann organizer formation(GO:0060061) |
| 0.1 | 0.9 | GO:0045899 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.1 | 0.3 | GO:0070655 | mechanosensory epithelium regeneration(GO:0070655) mechanoreceptor differentiation involved in mechanosensory epithelium regeneration(GO:0070656) neuromast regeneration(GO:0070657) neuromast hair cell differentiation involved in neuromast regeneration(GO:0070658) |
| 0.1 | 1.1 | GO:0051256 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) mitotic spindle midzone assembly(GO:0051256) |
| 0.1 | 0.3 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.1 | 3.9 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.1 | 0.5 | GO:0034205 | beta-amyloid formation(GO:0034205) amyloid precursor protein catabolic process(GO:0042987) |
| 0.1 | 0.8 | GO:0021754 | facial nucleus development(GO:0021754) |
| 0.1 | 1.0 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 2.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 2.0 | GO:0017144 | drug metabolic process(GO:0017144) drug catabolic process(GO:0042737) exogenous drug catabolic process(GO:0042738) |
| 0.1 | 0.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 0.5 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.6 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.8 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.9 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.1 | 0.3 | GO:0018872 | arsonoacetate metabolic process(GO:0018872) |
| 0.1 | 0.3 | GO:0032367 | intracellular cholesterol transport(GO:0032367) |
| 0.1 | 0.4 | GO:0071405 | response to methanol(GO:0033986) cellular response to methanol(GO:0071405) |
| 0.1 | 0.5 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 0.5 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.8 | GO:0070828 | heterochromatin organization(GO:0070828) |
| 0.1 | 0.3 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.1 | 0.3 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.1 | 3.5 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.5 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.0 | 0.3 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.0 | 0.2 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 1.9 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.8 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 1.1 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.4 | GO:0009113 | purine nucleobase biosynthetic process(GO:0009113) |
| 0.0 | 3.3 | GO:0001894 | tissue homeostasis(GO:0001894) |
| 0.0 | 0.6 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 1.0 | GO:0003094 | glomerular filtration(GO:0003094) |
| 0.0 | 1.3 | GO:0006367 | transcription initiation from RNA polymerase II promoter(GO:0006367) |
| 0.0 | 0.2 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 2.8 | GO:0090630 | activation of GTPase activity(GO:0090630) |
| 0.0 | 0.8 | GO:0035118 | embryonic pectoral fin morphogenesis(GO:0035118) |
| 0.0 | 0.6 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 1.0 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 1.1 | GO:0050821 | protein stabilization(GO:0050821) |
| 0.0 | 1.1 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.2 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.0 | 0.6 | GO:0006959 | humoral immune response(GO:0006959) |
| 0.0 | 0.3 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.2 | GO:1904825 | protein localization to microtubule(GO:0035372) protein localization to microtubule plus-end(GO:1904825) |
| 0.0 | 0.5 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 1.0 | GO:0030917 | midbrain-hindbrain boundary development(GO:0030917) |
| 0.0 | 0.4 | GO:0007492 | endoderm development(GO:0007492) |
| 0.0 | 0.4 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.2 | GO:0071910 | determination of pancreatic left/right asymmetry(GO:0035469) determination of liver left/right asymmetry(GO:0071910) |
| 0.0 | 0.1 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.3 | GO:0006406 | mRNA export from nucleus(GO:0006406) mRNA-containing ribonucleoprotein complex export from nucleus(GO:0071427) |
| 0.0 | 0.3 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) amino sugar biosynthetic process(GO:0046349) |
| 0.0 | 0.3 | GO:0031647 | regulation of protein stability(GO:0031647) |
| 0.0 | 0.1 | GO:0010915 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) regulation of lipoprotein particle clearance(GO:0010984) negative regulation of lipoprotein particle clearance(GO:0010985) |
| 0.0 | 0.3 | GO:0046660 | female sex differentiation(GO:0046660) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.6 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.4 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.0 | 0.2 | GO:0035794 | positive regulation of mitochondrial membrane permeability(GO:0035794) mitochondrial outer membrane permeabilization(GO:0097345) positive regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902110) mitochondrial outer membrane permeabilization involved in programmed cell death(GO:1902686) |
| 0.0 | 0.1 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 | 1.3 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 0.1 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
| 0.0 | 0.5 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.1 | GO:0051124 | postsynaptic membrane organization(GO:0001941) synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.4 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.8 | 3.1 | GO:0002144 | cytosolic tRNA wobble base thiouridylase complex(GO:0002144) |
| 0.3 | 1.3 | GO:0005674 | transcription factor TFIIF complex(GO:0005674) |
| 0.3 | 0.9 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.3 | 1.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 2.0 | GO:0005921 | gap junction(GO:0005921) |
| 0.2 | 1.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.2 | 1.9 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.2 | 0.7 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.2 | 3.7 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 1.2 | GO:0071546 | pi-body(GO:0071546) |
| 0.2 | 2.5 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.1 | 0.4 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.1 | 2.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 1.0 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.1 | 0.5 | GO:0031415 | NatA complex(GO:0031415) |
| 0.1 | 0.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.1 | 1.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.5 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 1.1 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.1 | 0.9 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 1.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 2.7 | GO:0005811 | lipid particle(GO:0005811) |
| 0.1 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.1 | 0.5 | GO:0089701 | U2AF(GO:0089701) |
| 0.1 | 0.8 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.1 | 0.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.1 | 0.8 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.4 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.1 | 2.0 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.1 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 3.7 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.1 | 0.7 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.1 | 0.6 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.1 | 1.7 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.6 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.6 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.7 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.5 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 1.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 7.8 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.2 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 1.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.0 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.1 | GO:0005753 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 1.8 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 2.6 | GO:0005768 | endosome(GO:0005768) |
| 0.0 | 0.2 | GO:0051233 | spindle midzone(GO:0051233) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.1 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.8 | 4.8 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.6 | 1.7 | GO:0030629 | U6 snRNA 3'-end binding(GO:0030629) 23S rRNA (adenine(1618)-N(6))-methyltransferase activity(GO:0052907) |
| 0.5 | 2.6 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.5 | 1.5 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.5 | 5.0 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.4 | 3.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.4 | 1.5 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.3 | 1.3 | GO:0003823 | antigen binding(GO:0003823) lipid antigen binding(GO:0030882) |
| 0.3 | 1.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.3 | 2.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.3 | 4.4 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.2 | 2.0 | GO:0022829 | wide pore channel activity(GO:0022829) |
| 0.2 | 0.6 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.2 | 0.8 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.2 | 1.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 0.7 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.2 | 0.7 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.2 | 0.5 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.2 | 3.0 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.1 | 0.4 | GO:0004044 | amidophosphoribosyltransferase activity(GO:0004044) |
| 0.1 | 0.9 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 0.6 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.1 | 2.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.9 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.1 | 0.4 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.3 | GO:0004475 | mannose-1-phosphate guanylyltransferase activity(GO:0004475) mannose-phosphate guanylyltransferase activity(GO:0008905) |
| 0.1 | 1.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 1.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.1 | 0.5 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.1 | 1.6 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 1.4 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 1.7 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.5 | GO:0046922 | peptide-O-fucosyltransferase activity(GO:0046922) |
| 0.1 | 0.3 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 0.3 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.1 | 1.0 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 0.4 | GO:0042910 | xenobiotic transporter activity(GO:0042910) |
| 0.1 | 1.7 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.1 | 1.7 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.1 | 0.6 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.1 | 3.1 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 5.0 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 0.8 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.1 | 0.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.1 | 0.3 | GO:0030792 | arsenite methyltransferase activity(GO:0030791) methylarsonite methyltransferase activity(GO:0030792) |
| 0.1 | 2.6 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.1 | 4.8 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.1 | 0.5 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.1 | 0.5 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) phosphoprotein binding(GO:0051219) |
| 0.1 | 3.6 | GO:0003724 | RNA helicase activity(GO:0003724) |
| 0.1 | 2.0 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.1 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.5 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 1.1 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0045134 | guanosine-diphosphatase activity(GO:0004382) uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 0.7 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.8 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 3.9 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.8 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.9 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.8 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 1.0 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 1.0 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 1.4 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 2.6 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.1 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 1.1 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.7 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) |
| 0.0 | 1.3 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.7 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.3 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 2.7 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.0 | 2.0 | GO:0019901 | protein kinase binding(GO:0019901) |
| 0.0 | 4.0 | GO:0008047 | enzyme activator activity(GO:0008047) |
| 0.0 | 0.5 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0004407 | histone deacetylase activity(GO:0004407) NAD-dependent histone deacetylase activity(GO:0017136) histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.0 | 3.0 | GO:0003729 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 3.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) collagen receptor activity(GO:0038064) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 3.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 2.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 1.3 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.1 | 1.9 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.1 | 1.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.1 | 0.5 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 3.2 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 2.0 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.4 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.8 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.1 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.2 | 3.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 2.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.7 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.1 | 0.5 | REACTOME SIGNALING BY NOTCH4 | Genes involved in Signaling by NOTCH4 |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 1.4 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 1.9 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.9 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.9 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.4 | REACTOME MRNA 3 END PROCESSING | Genes involved in mRNA 3'-end processing |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.7 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |