Project
DANIO-CODE
Navigation
Downloads
Results for tbx19
Z-value: 0.88
Transcription factors associated with tbx19
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbx19
|
ENSDARG00000079187 | T-box transcription factor 19 |
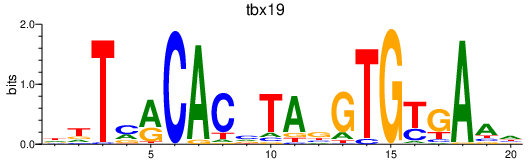
Activity profile of tbx19 motif
Sorted Z-values of tbx19 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of tbx19
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_32285237 | 2.62 |
ENSDART00000157324
|
prrg2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr15_+_29092022 | 2.24 |
ENSDART00000141164
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr9_+_8387050 | 2.21 |
ENSDART00000136847
|
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr17_+_15527923 | 2.02 |
ENSDART00000148443
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr17_-_20264870 | 1.97 |
|
|
|
| chr17_+_23291331 | 1.96 |
ENSDART00000128073
|
ppp1r3ca
|
protein phosphatase 1, regulatory subunit 3Ca |
| chr15_+_29092224 | 1.93 |
ENSDART00000131755
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr14_+_34150130 | 1.89 |
ENSDART00000132193
ENSDART00000141058 |
wnt8a
BX927327.1
|
wingless-type MMTV integration site family, member 8a ENSDARG00000105311 |
| chr17_-_20264762 | 1.85 |
|
|
|
| chr5_-_31173320 | 1.60 |
ENSDART00000122066
|
fam102ab
|
family with sequence similarity 102, member Ab |
| KN150339v1_-_39357 | 1.59 |
ENSDART00000169638
|
wu:fa19b12
|
wu:fa19b12 |
| chr18_+_47305642 | 1.56 |
ENSDART00000009775
|
rbm7
|
RNA binding motif protein 7 |
| chr11_-_11809024 | 1.54 |
ENSDART00000011657
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr10_+_15644868 | 1.51 |
ENSDART00000139259
|
zfand5b
|
zinc finger, AN1-type domain 5b |
| chr15_+_29091983 | 1.46 |
ENSDART00000141164
|
si:ch211-137a8.2
|
si:ch211-137a8.2 |
| chr15_-_25333773 | 1.44 |
ENSDART00000078230
|
mettl16
|
methyltransferase like 16 |
| chr6_-_8156471 | 1.38 |
ENSDART00000081561
|
ilf3a
|
interleukin enhancer binding factor 3a |
| chr1_-_18118467 | 1.38 |
ENSDART00000142026
|
si:dkey-167i21.2
|
si:dkey-167i21.2 |
| chr5_+_24039736 | 1.35 |
ENSDART00000175389
|
atp6v0a2b
|
ATPase, H+ transporting, lysosomal V0 subunit a2b |
| chr5_-_23211957 | 1.34 |
ENSDART00000019992
|
gbgt1l1
|
globoside alpha-1,3-N-acetylgalactosaminyltransferase 1, like 1 |
| chr5_+_29052293 | 1.24 |
ENSDART00000035400
|
tsc1a
|
tuberous sclerosis 1a |
| chr4_+_50616647 | 1.23 |
|
|
|
| KN150712v1_-_26150 | 1.19 |
|
|
|
| chr6_+_4185188 | 1.18 |
|
|
|
| chr13_+_8364704 | 1.13 |
ENSDART00000109059
|
ttc7a
|
tetratricopeptide repeat domain 7A |
| chr17_+_15527861 | 1.12 |
ENSDART00000148443
|
marcksb
|
myristoylated alanine-rich protein kinase C substrate b |
| chr15_+_45590082 | 1.07 |
|
|
|
| chr20_+_7337986 | 1.07 |
ENSDART00000140222
|
dsg2.1
|
desmoglein 2, tandem duplicate 1 |
| chr6_-_22842200 | 1.06 |
ENSDART00000160311
|
sumo2a
|
small ubiquitin-like modifier 2a |
| chr17_+_34238753 | 1.04 |
ENSDART00000014306
|
mpp5a
|
membrane protein, palmitoylated 5a (MAGUK p55 subfamily member 5) |
| chr23_+_20614283 | 1.00 |
|
|
|
| chr8_+_10824790 | 0.96 |
ENSDART00000140717
|
brpf3b
|
bromodomain and PHD finger containing, 3b |
| chr6_-_39634534 | 0.95 |
ENSDART00000077839
|
atf7b
|
activating transcription factor 7b |
| chr14_-_6220150 | 0.95 |
ENSDART00000125058
|
nipsnap3a
|
nipsnap homolog 3A (C. elegans) |
| chr5_-_32290516 | 0.91 |
ENSDART00000007512
|
pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr3_-_24923814 | 0.89 |
ENSDART00000086768
|
ep300b
|
E1A binding protein p300 b |
| chr5_-_7206937 | 0.89 |
ENSDART00000168820
|
bmpr1ba
|
bone morphogenetic protein receptor, type IBa |
| chr5_-_34664282 | 0.89 |
ENSDART00000114981
ENSDART00000051279 |
ptcd2
|
pentatricopeptide repeat domain 2 |
| chr9_+_27909662 | 0.89 |
ENSDART00000112415
|
lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr11_+_12802775 | 0.88 |
ENSDART00000037474
|
zgpat
|
zinc finger, CCCH-type with G patch domain |
| chr5_-_24039681 | 0.87 |
ENSDART00000046384
|
trmt2a
|
tRNA methyltransferase 2 homolog A |
| chr5_+_29052517 | 0.86 |
ENSDART00000035400
|
tsc1a
|
tuberous sclerosis 1a |
| chr9_+_38832959 | 0.86 |
ENSDART00000110651
|
slc12a8
|
solute carrier family 12, member 8 |
| chr9_-_49957348 | 0.85 |
|
|
|
| chr9_-_49957319 | 0.84 |
|
|
|
| chr4_-_29095531 | 0.83 |
ENSDART00000142951
|
si:dkey-23a23.2
|
si:dkey-23a23.2 |
| chr13_-_44694139 | 0.81 |
ENSDART00000137275
|
si:dkeyp-2e4.6
|
si:dkeyp-2e4.6 |
| chr15_-_25333252 | 0.80 |
ENSDART00000127771
|
mettl16
|
methyltransferase like 16 |
| chr6_+_41506681 | 0.80 |
ENSDART00000084861
|
cish
|
cytokine inducible SH2-containing protein |
| KN150702v1_-_111659 | 0.79 |
ENSDART00000165431
|
PPP1CC
|
protein phosphatase 1 catalytic subunit gamma |
| chr2_-_31817448 | 0.79 |
ENSDART00000170880
|
retreg1
|
reticulophagy regulator 1 |
| chr13_-_44671682 | 0.78 |
ENSDART00000099984
|
glo1
|
glyoxalase 1 |
| chr18_+_39125694 | 0.78 |
ENSDART00000122377
|
bcl2l10
|
BCL2-like 10 (apoptosis facilitator) |
| chr12_-_26939563 | 0.75 |
ENSDART00000076103
|
bc2
|
putative breast adenocarcinoma marker |
| chr24_-_17001295 | 0.74 |
|
|
|
| chr8_-_18192956 | 0.74 |
ENSDART00000110249
|
si:ch211-241d21.5
|
si:ch211-241d21.5 |
| chr2_-_31817409 | 0.74 |
ENSDART00000112763
|
retreg1
|
reticulophagy regulator 1 |
| chr8_-_38284904 | 0.73 |
ENSDART00000125920
|
pdlim2
|
PDZ and LIM domain 2 (mystique) |
| chr8_+_20383989 | 0.72 |
ENSDART00000009081
|
mob3a
|
MOB kinase activator 3A |
| chr5_+_55607591 | 0.71 |
ENSDART00000148749
|
aatf
|
apoptosis antagonizing transcription factor |
| chr9_-_41222630 | 0.70 |
ENSDART00000157398
|
pms1
|
PMS1 homolog 1, mismatch repair system component |
| chr2_+_25212281 | 0.70 |
ENSDART00000078838
|
rab3aa
|
RAB3A, member RAS oncogene family, a |
| chr5_+_29052462 | 0.70 |
ENSDART00000035400
|
tsc1a
|
tuberous sclerosis 1a |
| chr21_+_34942142 | 0.69 |
ENSDART00000139635
|
rbm11
|
RNA binding motif protein 11 |
| chr23_-_31986679 | 0.68 |
ENSDART00000085054
|
mtfr2
|
mitochondrial fission regulator 2 |
| chr9_-_55539103 | 0.67 |
ENSDART00000085135
|
tbl1x
|
transducin (beta)-like 1X-linked |
| chr5_-_54578223 | 0.67 |
ENSDART00000016995
ENSDART00000134937 |
srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr17_-_50411385 | 0.67 |
ENSDART00000075159
|
zgc:113377
|
zgc:113377 |
| chr4_-_19039862 | 0.66 |
ENSDART00000166374
|
si:dkey-31f5.11
|
si:dkey-31f5.11 |
| chr5_-_32290272 | 0.66 |
ENSDART00000007512
|
pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr12_-_33704092 | 0.65 |
ENSDART00000030566
|
galk1
|
galactokinase 1 |
| chr1_+_18118735 | 0.65 |
ENSDART00000078610
|
slc25a51a
|
solute carrier family 25, member 51a |
| chr24_+_25995817 | 0.62 |
ENSDART00000139017
|
tmtopsb
|
teleost multiple tissue opsin b |
| chr3_+_24167185 | 0.60 |
ENSDART00000154078
|
CR812472.2
|
ENSDARG00000097380 |
| chr6_+_40525779 | 0.60 |
ENSDART00000033819
|
prkcda
|
protein kinase C, delta a |
| chr23_-_3778530 | 0.59 |
ENSDART00000141682
|
nudt3a
|
nudix (nucleoside diphosphate linked moiety X)-type motif 3a |
| chr5_-_32290231 | 0.57 |
ENSDART00000007512
|
pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr6_-_39633603 | 0.57 |
ENSDART00000104042
|
atf7b
|
activating transcription factor 7b |
| chr10_-_33399226 | 0.55 |
ENSDART00000164191
|
c2cd2
|
C2 calcium-dependent domain containing 2 |
| chr1_+_19071918 | 0.54 |
ENSDART00000138276
|
ENSDARG00000077648
|
ENSDARG00000077648 |
| chr6_+_19933322 | 0.54 |
ENSDART00000143378
|
thumpd3
|
THUMP domain containing 3 |
| chr24_+_31751950 | 0.52 |
ENSDART00000156493
|
CT737162.1
|
ENSDARG00000096998 |
| chr3_+_38939974 | 0.52 |
|
|
|
| chr17_-_33754959 | 0.51 |
ENSDART00000124788
|
si:dkey-84k17.2
|
si:dkey-84k17.2 |
| chr4_+_13490875 | 0.51 |
ENSDART00000178376
|
BX465210.1
|
ENSDARG00000107553 |
| chr9_-_27909272 | 0.50 |
ENSDART00000161068
|
gtf2e1
|
general transcription factor IIE, polypeptide 1, alpha |
| chr10_+_6381994 | 0.50 |
ENSDART00000170548
|
ENSDARG00000100633
|
ENSDARG00000100633 |
| chr18_+_16144493 | 0.50 |
ENSDART00000080423
|
ctsd
|
cathepsin D |
| chr10_+_5267937 | 0.49 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr9_+_8386966 | 0.49 |
ENSDART00000138713
|
si:dkey-90l23.2
|
si:dkey-90l23.2 |
| chr3_+_32285593 | 0.48 |
ENSDART00000156986
|
prrg2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr17_-_50377407 | 0.47 |
ENSDART00000156110
|
si:ch211-235i11.7
|
si:ch211-235i11.7 |
| chr22_+_12570345 | 0.46 |
ENSDART00000140054
|
ENSDARG00000041602
|
ENSDARG00000041602 |
| chr16_+_1843899 | 0.46 |
|
|
|
| chr12_-_28841710 | 0.46 |
ENSDART00000142933
|
znf646
|
zinc finger protein 646 |
| chr17_-_24872401 | 0.44 |
ENSDART00000135569
|
gale
|
UDP-galactose-4-epimerase |
| chr14_-_50657712 | 0.44 |
ENSDART00000169456
|
nfkb1
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| KN150702v1_-_111618 | 0.44 |
ENSDART00000165431
|
PPP1CC
|
protein phosphatase 1 catalytic subunit gamma |
| chr5_-_7206860 | 0.43 |
ENSDART00000168820
|
bmpr1ba
|
bone morphogenetic protein receptor, type IBa |
| chr14_+_34150232 | 0.42 |
ENSDART00000148044
|
wnt8a
|
wingless-type MMTV integration site family, member 8a |
| chr12_+_10078043 | 0.42 |
ENSDART00000152369
|
si:dkeyp-118b1.2
|
si:dkeyp-118b1.2 |
| chr25_+_28636948 | 0.41 |
|
|
|
| chr5_-_32290543 | 0.40 |
ENSDART00000007512
|
pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr14_+_28180174 | 0.40 |
ENSDART00000161852
|
stag2b
|
stromal antigen 2b |
| chr20_-_38851375 | 0.39 |
|
|
|
| chr7_-_19851755 | 0.39 |
ENSDART00000052902
|
acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
| chr5_-_54578261 | 0.38 |
ENSDART00000016995
ENSDART00000134937 |
srek1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr9_+_27909514 | 0.37 |
ENSDART00000144481
|
lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr20_+_38934332 | 0.37 |
ENSDART00000061334
|
ift172
|
intraflagellar transport 172 |
| chr8_-_18193491 | 0.37 |
ENSDART00000143036
|
si:ch211-241d21.5
|
si:ch211-241d21.5 |
| chr10_+_5268009 | 0.37 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr15_-_92920 | 0.37 |
ENSDART00000099074
|
si:zfos-411a11.2
|
si:zfos-411a11.2 |
| chr15_-_25333737 | 0.36 |
ENSDART00000078230
|
mettl16
|
methyltransferase like 16 |
| chr25_+_17906515 | 0.36 |
|
|
|
| chr5_-_31173039 | 0.35 |
ENSDART00000137556
|
fam102ab
|
family with sequence similarity 102, member Ab |
| chr18_+_40365208 | 0.34 |
ENSDART00000167134
|
sema6dl
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D, like |
| chr20_+_9136578 | 0.34 |
ENSDART00000064150
ENSDART00000158196 |
ENSDARG00000043687
|
ENSDARG00000043687 |
| chr3_+_32285564 | 0.33 |
ENSDART00000156986
|
prrg2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr17_+_50310712 | 0.32 |
ENSDART00000154246
|
znf982
|
zinc finger protein 982 |
| chr13_-_35716712 | 0.32 |
ENSDART00000170106
|
map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr9_+_21042980 | 0.31 |
ENSDART00000003648
|
wdr3
|
WD repeat domain 3 |
| chr16_+_33948113 | 0.31 |
ENSDART00000159474
|
gnl2
|
guanine nucleotide binding protein-like 2 (nucleolar) |
| chr1_-_19072200 | 0.31 |
|
|
|
| chr5_+_31475897 | 0.31 |
ENSDART00000144510
|
zmat5
|
zinc finger, matrin-type 5 |
| chr11_-_11808973 | 0.30 |
ENSDART00000011657
|
cdc6
|
cell division cycle 6 homolog (S. cerevisiae) |
| chr3_+_40022244 | 0.29 |
ENSDART00000102526
|
gfer
|
growth factor, augmenter of liver regeneration (ERV1 homolog, S. cerevisiae) |
| chr6_-_8156520 | 0.29 |
ENSDART00000081561
|
ilf3a
|
interleukin enhancer binding factor 3a |
| chr5_+_35355253 | 0.29 |
ENSDART00000074630
|
zgc:103697
|
zgc:103697 |
| chr10_+_22538287 | 0.27 |
ENSDART00000109070
|
gigyf1b
|
GRB10 interacting GYF protein 1b |
| chr22_-_18519706 | 0.27 |
ENSDART00000132156
|
cirbpb
|
cold inducible RNA binding protein b |
| chr14_-_49951804 | 0.26 |
ENSDART00000021736
|
scocb
|
short coiled-coil protein b |
| chr6_-_40654280 | 0.26 |
|
|
|
| chr24_+_17001576 | 0.25 |
ENSDART00000149744
|
mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr24_+_14070379 | 0.24 |
ENSDART00000004664
|
tram1
|
translocation associated membrane protein 1 |
| chr7_-_6232554 | 0.24 |
ENSDART00000127042
|
HIST2H2AB (1 of many)
|
histone cluster 2 H2A family member b |
| chr24_+_17001271 | 0.23 |
ENSDART00000149744
|
mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr5_-_1734159 | 0.23 |
ENSDART00000145781
|
ENSDARG00000094300
|
ENSDARG00000094300 |
| chr22_+_8286249 | 0.22 |
ENSDART00000103911
|
CABZ01077217.1
|
ENSDARG00000079145 |
| chr4_-_71770315 | 0.21 |
ENSDART00000171434
|
zgc:162958
|
zgc:162958 |
| chr13_+_40560230 | 0.21 |
ENSDART00000146112
|
hps1
|
Hermansky-Pudlak syndrome 1 |
| chr7_-_33610248 | 0.21 |
ENSDART00000100102
|
rxfp3.3a1
|
relaxin/insulin-like family peptide receptor 3.3a1 |
| chr4_-_29151687 | 0.21 |
ENSDART00000176602
|
CR388132.4
|
ENSDARG00000106707 |
| chr19_+_32614657 | 0.21 |
ENSDART00000022667
|
fam8a1a
|
family with sequence similarity 8, member A1a |
| chr11_+_24562450 | 0.21 |
ENSDART00000082761
ENSDART00000131976 |
adipor1a
|
adiponectin receptor 1a |
| chr10_+_5267746 | 0.20 |
ENSDART00000114491
|
ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr7_-_38299706 | 0.20 |
ENSDART00000041055
|
celf1
|
cugbp, Elav-like family member 1 |
| chr7_+_32880600 | 0.19 |
ENSDART00000029106
|
si:ch211-194p6.10
|
si:ch211-194p6.10 |
| chr17_+_14776302 | 0.18 |
ENSDART00000154229
|
RTRAF
|
zgc:56576 |
| chr15_+_45589989 | 0.17 |
|
|
|
| chr20_+_19094480 | 0.16 |
ENSDART00000152380
|
tdh
|
L-threonine dehydrogenase |
| chr6_+_54798741 | 0.16 |
|
|
|
| chr12_-_15524614 | 0.16 |
ENSDART00000053003
|
hexim1
|
hexamethylene bis-acetamide inducible 1 |
| chr7_-_71214600 | 0.16 |
ENSDART00000042492
|
sepsecs
|
Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| chr22_-_18520332 | 0.15 |
|
|
|
| chr8_-_43151611 | 0.15 |
ENSDART00000134801
|
ccdc92
|
coiled-coil domain containing 92 |
| chr7_-_38299544 | 0.15 |
ENSDART00000041055
|
celf1
|
cugbp, Elav-like family member 1 |
| chr9_+_32367808 | 0.15 |
ENSDART00000078576
|
coq10b
|
coenzyme Q10B |
| chr17_-_20147162 | 0.14 |
ENSDART00000104874
|
p4ha1b
|
prolyl 4-hydroxylase, alpha polypeptide I b |
| chr6_+_40525610 | 0.14 |
ENSDART00000033819
|
prkcda
|
protein kinase C, delta a |
| chr11_-_15981084 | 0.13 |
ENSDART00000164215
|
rpf1
|
ribosome production factor 1 homolog |
| chr4_+_38593055 | 0.11 |
ENSDART00000150397
|
znf977
|
zinc finger protein 977 |
| chr10_-_28474052 | 0.11 |
|
|
|
| chr4_-_62271066 | 0.09 |
ENSDART00000162955
|
znf1091
|
zinc finger protein 1091 |
| chr4_-_64349492 | 0.08 |
|
|
|
| chr3_+_36830369 | 0.07 |
ENSDART00000150917
|
ENSDARG00000070174
|
ENSDARG00000070174 |
| chr25_-_17906739 | 0.06 |
ENSDART00000149696
|
cep290
|
centrosomal protein 290 |
| chr22_+_4563528 | 0.02 |
ENSDART00000122324
|
si:ch1073-104i17.1
|
si:ch1073-104i17.1 |
| chr21_-_20295921 | 0.02 |
ENSDART00000166049
|
slc26a1
|
solute carrier family 26 (anion exchanger), member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:1900144 | epithelial cell morphogenesis involved in gastrulation(GO:0003381) BMP secretion(GO:0038055) positive regulation of BMP secretion(GO:1900144) regulation of BMP secretion(GO:2001284) |
| 0.6 | 2.3 | GO:0021512 | spinal cord anterior/posterior patterning(GO:0021512) cell proliferation involved in pronephros development(GO:0039015) cell proliferation involved in kidney development(GO:0072111) |
| 0.5 | 2.5 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.3 | 1.3 | GO:0031591 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.3 | 1.8 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.3 | 1.6 | GO:0021693 | cerebellar Purkinje cell layer structural organization(GO:0021693) cerebellar cortex structural organization(GO:0021698) |
| 0.2 | 2.8 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.2 | 0.9 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.2 | 2.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.2 | 0.9 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.2 | 0.7 | GO:0099558 | maintenance of presynaptic active zone structure(GO:0048790) maintenance of synapse structure(GO:0099558) |
| 0.1 | 2.0 | GO:0010962 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glucan biosynthetic process(GO:0010962) |
| 0.1 | 1.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 1.4 | GO:0045851 | vacuolar acidification(GO:0007035) pH reduction(GO:0045851) intracellular pH reduction(GO:0051452) |
| 0.1 | 1.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.1 | 0.3 | GO:0061031 | endodermal digestive tract morphogenesis(GO:0061031) |
| 0.1 | 0.8 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.1 | 1.4 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.1 | 0.5 | GO:0034367 | macromolecular complex remodeling(GO:0034367) |
| 0.1 | 0.7 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 1.0 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.1 | 0.6 | GO:1901909 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.1 | 0.8 | GO:0010522 | regulation of calcium ion transport into cytosol(GO:0010522) regulation of release of sequestered calcium ion into cytosol(GO:0051279) |
| 0.1 | 0.2 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.9 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.1 | 1.3 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.1 | 1.7 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 0.2 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.1 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.5 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 1.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.1 | GO:0014068 | positive regulation of phosphatidylinositol 3-kinase signaling(GO:0014068) |
| 0.0 | 0.7 | GO:0006298 | mismatch repair(GO:0006298) |
| 0.0 | 0.2 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.2 | GO:0006566 | threonine metabolic process(GO:0006566) threonine catabolic process(GO:0006567) |
| 0.0 | 0.7 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.5 | GO:0060729 | maintenance of gastrointestinal epithelium(GO:0030277) intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 1.7 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.2 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 1.1 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.9 | GO:0050684 | regulation of mRNA processing(GO:0050684) |
| 0.0 | 0.9 | GO:0033339 | pectoral fin development(GO:0033339) |
| 0.0 | 0.4 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) |
| 0.0 | 0.7 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.0 | 0.4 | GO:0007062 | sister chromatid cohesion(GO:0007062) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.6 | 2.3 | GO:0097189 | apoptotic body(GO:0097189) |
| 0.5 | 2.5 | GO:0008623 | CHRAC(GO:0008623) |
| 0.4 | 5.6 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.2 | 0.9 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.2 | 1.4 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.1 | 2.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.7 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.1 | 0.2 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 3.8 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.1 | 1.0 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.1 | 0.6 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.0 | 0.4 | GO:0008278 | cohesin complex(GO:0008278) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.3 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0045495 | pole plasm(GO:0045495) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0052907 | U6 snRNA 3'-end binding(GO:0030629) 23S rRNA (adenine(1618)-N(6))-methyltransferase activity(GO:0052907) |
| 0.1 | 2.0 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 1.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.1 | 0.7 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.1 | 1.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.8 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 1.0 | GO:0043994 | H3 histone acetyltransferase activity(GO:0010484) histone acetyltransferase activity (H3-K23 specific)(GO:0043994) |
| 0.1 | 0.6 | GO:0008486 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.1 | 1.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.1 | 0.9 | GO:0015379 | cation:chloride symporter activity(GO:0015377) potassium:chloride symporter activity(GO:0015379) |
| 0.1 | 1.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 0.7 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 3.7 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.1 | 2.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.1 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 0.2 | GO:0008743 | L-threonine 3-dehydrogenase activity(GO:0008743) |
| 0.1 | 1.1 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.3 | GO:0016972 | flavin-linked sulfhydryl oxidase activity(GO:0016971) thiol oxidase activity(GO:0016972) |
| 0.0 | 1.8 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.7 | GO:0004698 | protein kinase C activity(GO:0004697) calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 3.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.5 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.7 | GO:0051393 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.9 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.0 | 1.1 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 2.7 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.5 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 1.6 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 1.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.8 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 1.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 2.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 1.8 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.8 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.7 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.5 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |