Project
DANIO-CODE
Navigation
Downloads
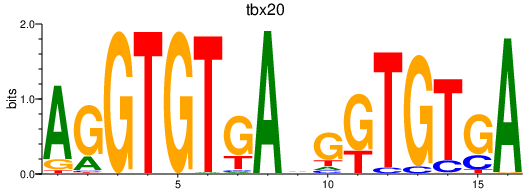
Results for tbx20
Z-value: 0.38
Transcription factors associated with tbx20
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
tbx20
|
ENSDARG00000005150 | T-box transcription factor 20 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| tbx20 | dr10_dc_chr16_-_42619967_42619993 | -0.24 | 3.7e-01 | Click! |
Activity profile of tbx20 motif
Sorted Z-values of tbx20 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of tbx20
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_42761097 | 1.03 |
ENSDART00000075259
|
fgfr1b
|
fibroblast growth factor receptor 1b |
| chr12_-_43917936 | 0.63 |
ENSDART00000159423
|
foxi1
|
forkhead box i1 |
| chr23_+_43785707 | 0.47 |
ENSDART00000172160
|
ENSDARG00000105093
|
ENSDARG00000105093 |
| chr17_+_26946957 | 0.44 |
ENSDART00000114215
|
grhl3
|
grainyhead-like transcription factor 3 |
| chr17_-_25865861 | 0.43 |
|
|
|
| chr17_+_33422832 | 0.40 |
ENSDART00000032827
|
zgc:162964
|
zgc:162964 |
| chr10_+_42761028 | 0.39 |
ENSDART00000075259
|
fgfr1b
|
fibroblast growth factor receptor 1b |
| chr17_+_33423057 | 0.38 |
ENSDART00000032827
|
zgc:162964
|
zgc:162964 |
| chr5_-_45219030 | 0.35 |
|
|
|
| chr22_-_36721626 | 0.32 |
ENSDART00000017188
ENSDART00000124698 |
ncl
|
nucleolin |
| chr10_+_42761437 | 0.29 |
ENSDART00000075269
ENSDART00000067689 |
fgfr1b
|
fibroblast growth factor receptor 1b |
| chr2_+_23600656 | 0.29 |
|
|
|
| chr11_+_44360399 | 0.28 |
ENSDART00000159068
|
rbm34
|
RNA binding motif protein 34 |
| chr24_+_43010 | 0.25 |
ENSDART00000122785
|
zgc:152808
|
zgc:152808 |
| chr15_-_41732512 | 0.25 |
ENSDART00000059327
|
spsb4b
|
splA/ryanodine receptor domain and SOCS box containing 4b |
| chr3_-_6078015 | 0.24 |
ENSDART00000165715
|
BX284638.1
|
ENSDARG00000098850 |
| chr21_+_1997858 | 0.24 |
|
|
|
| chr21_-_1956120 | 0.24 |
ENSDART00000165547
|
CABZ01045062.1
|
ENSDARG00000100274 |
| chr5_+_30159040 | 0.22 |
ENSDART00000078018
|
ruvbl2
|
RuvB-like AAA ATPase 2 |
| chr12_+_4885394 | 0.21 |
ENSDART00000159367
|
plekhm1
|
pleckstrin homology domain containing, family M (with RUN domain) member 1 |
| chr23_+_43785682 | 0.19 |
ENSDART00000172160
|
ENSDARG00000105093
|
ENSDARG00000105093 |
| chr12_+_48356793 | 0.18 |
ENSDART00000054788
ENSDART00000152899 |
ddit4
|
DNA-damage-inducible transcript 4 |
| chr7_+_34482282 | 0.17 |
|
|
|
| chr20_+_42349759 | 0.17 |
ENSDART00000160519
ENSDART00000165631 |
gopc
|
golgi-associated PDZ and coiled-coil motif containing |
| chr15_-_31635737 | 0.16 |
ENSDART00000156047
|
hmgb1b
|
high mobility group box 1b |
| chr22_-_37861903 | 0.15 |
ENSDART00000085931
|
acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr12_+_1286911 | 0.14 |
ENSDART00000168225
ENSDART00000157467 |
pemt
|
phosphatidylethanolamine N-methyltransferase |
| chr12_-_8780098 | 0.14 |
ENSDART00000113148
|
jmjd1cb
|
jumonji domain containing 1Cb |
| chr18_+_5707331 | 0.14 |
|
|
|
| chr2_+_43620000 | 0.13 |
ENSDART00000142078
ENSDART00000098265 ENSDART00000098267 |
nrp1b
|
neuropilin 1b |
| chr21_+_2728070 | 0.12 |
|
|
|
| chr3_+_25726394 | 0.12 |
ENSDART00000097679
|
zgc:171844
|
zgc:171844 |
| chr13_-_10488076 | 0.12 |
ENSDART00000160561
|
si:ch73-54n14.2
|
si:ch73-54n14.2 |
| chr18_-_39476503 | 0.12 |
|
|
|
| chr2_+_1596524 | 0.12 |
ENSDART00000163310
|
exosc4
|
exosome component 4 |
| chr18_-_782643 | 0.11 |
ENSDART00000161084
|
si:dkey-205h23.1
|
si:dkey-205h23.1 |
| chr25_+_34602991 | 0.10 |
|
|
|
| chr14_+_20614087 | 0.10 |
ENSDART00000160318
|
lygl2
|
lysozyme g-like 2 |
| chr20_-_15181042 | 0.09 |
ENSDART00000175585
|
prrc2c
|
proline-rich coiled-coil 2C |
| chr2_+_43619953 | 0.09 |
ENSDART00000142078
ENSDART00000098265 ENSDART00000098267 |
nrp1b
|
neuropilin 1b |
| chr17_+_26946875 | 0.08 |
ENSDART00000114215
|
grhl3
|
grainyhead-like transcription factor 3 |
| chr24_+_42964 | 0.08 |
ENSDART00000122785
|
zgc:152808
|
zgc:152808 |
| chr23_-_43785497 | 0.08 |
ENSDART00000165963
|
ENSDARG00000102050
|
ENSDARG00000102050 |
| chr3_+_24587720 | 0.07 |
|
|
|
| chr13_+_10488748 | 0.06 |
ENSDART00000008603
|
prepl
|
prolyl endopeptidase-like |
| chr1_-_467033 | 0.06 |
ENSDART00000152799
|
ENSDARG00000078878
|
ENSDARG00000078878 |
| chr24_-_41356844 | 0.06 |
ENSDART00000121592
ENSDART00000019975 ENSDART00000166034 |
acvr2b
|
activin A receptor, type IIB |
| chr12_+_44772179 | 0.06 |
ENSDART00000112750
|
dhx32a
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32a |
| chr6_+_20595096 | 0.04 |
|
|
|
| chr22_-_37861837 | 0.04 |
ENSDART00000124742
|
acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr24_-_41356967 | 0.04 |
ENSDART00000121592
ENSDART00000019975 ENSDART00000166034 |
acvr2b
|
activin A receptor, type IIB |
| chr10_+_30771 | 0.03 |
ENSDART00000040240
|
tiprl
|
TIP41, TOR signaling pathway regulator-like (S. cerevisiae) |
| chr7_+_34482202 | 0.02 |
|
|
|
| chr13_+_34563757 | 0.02 |
ENSDART00000133661
|
tasp1
|
taspase, threonine aspartase, 1 |
| chr2_+_43619752 | 0.02 |
ENSDART00000142078
ENSDART00000098265 ENSDART00000098267 |
nrp1b
|
neuropilin 1b |
| chr25_-_7526393 | 0.02 |
|
|
|
| KN150135v1_+_3896 | 0.02 |
ENSDART00000174906
|
CABZ01008508.1
|
ENSDARG00000106459 |
| chr22_-_36721592 | 0.02 |
ENSDART00000130171
|
ncl
|
nucleolin |
| chr16_+_6809981 | 0.02 |
ENSDART00000149720
|
znf236
|
zinc finger protein 236 |
| chr7_+_37978605 | 0.02 |
ENSDART00000150158
|
si:dkey-78a14.4
|
si:dkey-78a14.4 |
| chr24_+_42884 | 0.01 |
ENSDART00000122785
|
zgc:152808
|
zgc:152808 |
| chr4_-_18851954 | 0.01 |
ENSDART00000140722
|
cbll1
|
Cbl proto-oncogene-like 1, E3 ubiquitin protein ligase |
| chr20_+_31314629 | 0.01 |
ENSDART00000020252
|
pdia6
|
protein disulfide isomerase family A, member 6 |
| chr11_-_12121214 | 0.01 |
ENSDART00000133236
ENSDART00000104226 |
mrpl45
|
mitochondrial ribosomal protein L45 |
| chr11_-_12121242 | 0.01 |
ENSDART00000136108
|
mrpl45
|
mitochondrial ribosomal protein L45 |
| chr11_-_44360143 | 0.01 |
ENSDART00000173340
|
tbce
|
tubulin folding cofactor E |
| chr4_-_18851997 | 0.01 |
ENSDART00000140722
|
cbll1
|
Cbl proto-oncogene-like 1, E3 ubiquitin protein ligase |
| chr24_-_41357096 | 0.00 |
ENSDART00000150207
|
acvr2b
|
activin A receptor, type IIB |
| chr13_+_30401664 | 0.00 |
|
|
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 0.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 1.7 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.2 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) negative regulation of protein localization to cell surface(GO:2000009) |
| 0.0 | 0.2 | GO:0035767 | endothelial cell chemotaxis(GO:0035767) |
| 0.0 | 0.2 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.1 | GO:0071554 | cell wall macromolecule metabolic process(GO:0044036) cell wall organization or biogenesis(GO:0071554) |
| 0.0 | 0.3 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.2 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.8 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.2 | PID ARF6 PATHWAY | Arf6 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |